This guide to the modules on the Tissue page is illustrated in the Common Metabolic Diseases Knowledge Portal (CMDKP) but applies to all Knowledge Portals that display genetic and genomic results.
The in-development Tissue page presents results specific to a particular tissue. Currently, it includes gene expression and global enrichment results. Please contact us with your suggestions on data types you would like to see on this page!
Begin new search
The Begin new search box at the top of the Tissue page allows you to navigate to a different Gene, Variant, Region, Phenotype, or Tissue page. Begin typing in the box and select an option from the list.
Gene expression section
This section displays absolute expression levels and tissue specificity of expression in the tissue whose page you are viewing. Currently, gene expression data are available for these tissues.
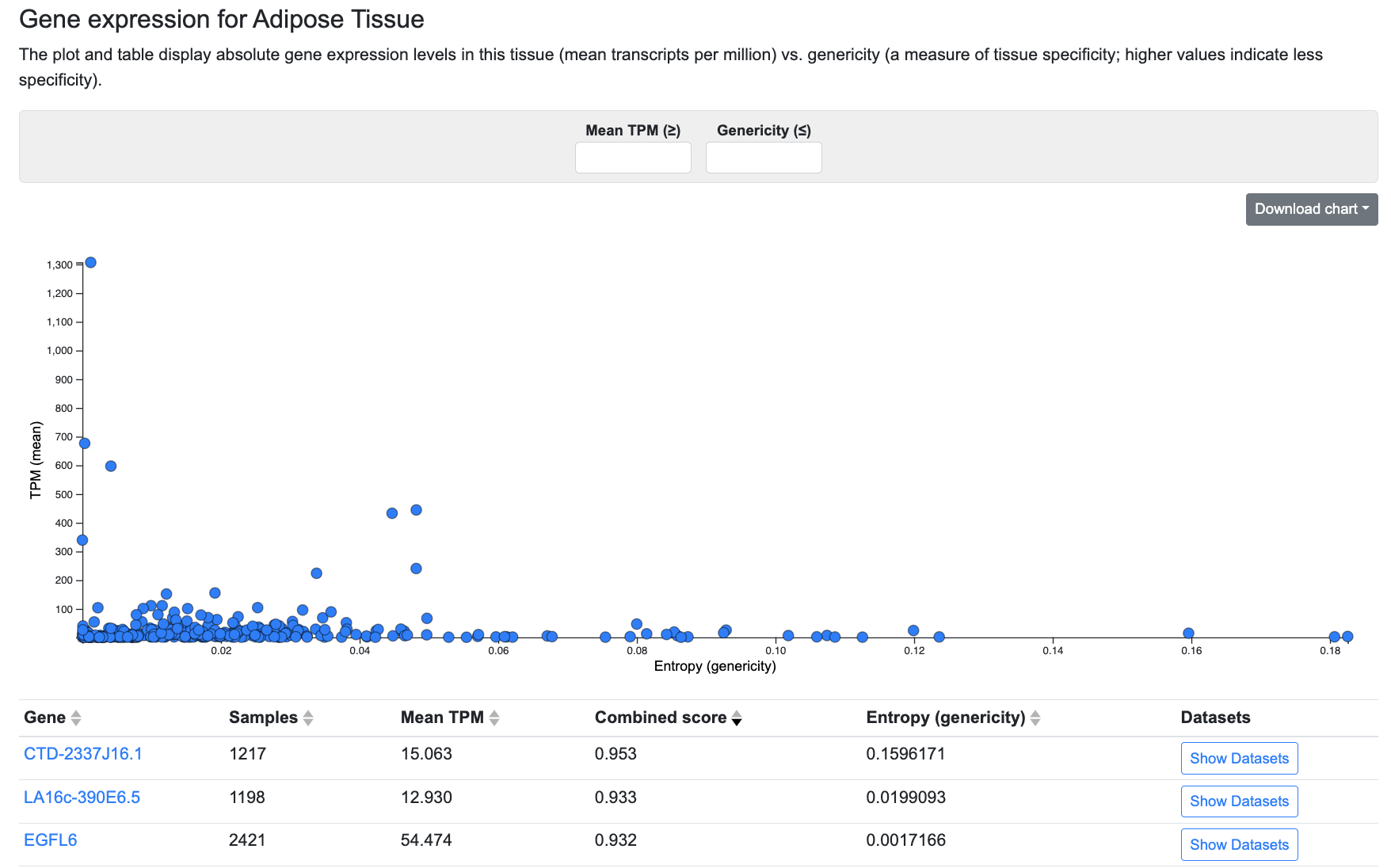
In the plot, the y axis represents absolute gene expression levels, in transcripts per million (TPM), while the x axis represents entropy or genericity, a measure of the tissue-specificity of gene expression. Low genericity indicates that expression of a gene is highly specific to the tissue whose page you are viewing, while high genericity indicates that a gene is expressed in many tissues. Mouse over the plot to see information about each gene.
The menus above the plot allow you to filter the plot and table by setting a threshold for mean TPM and/or genericity.
Results are also displayed in tabular format:
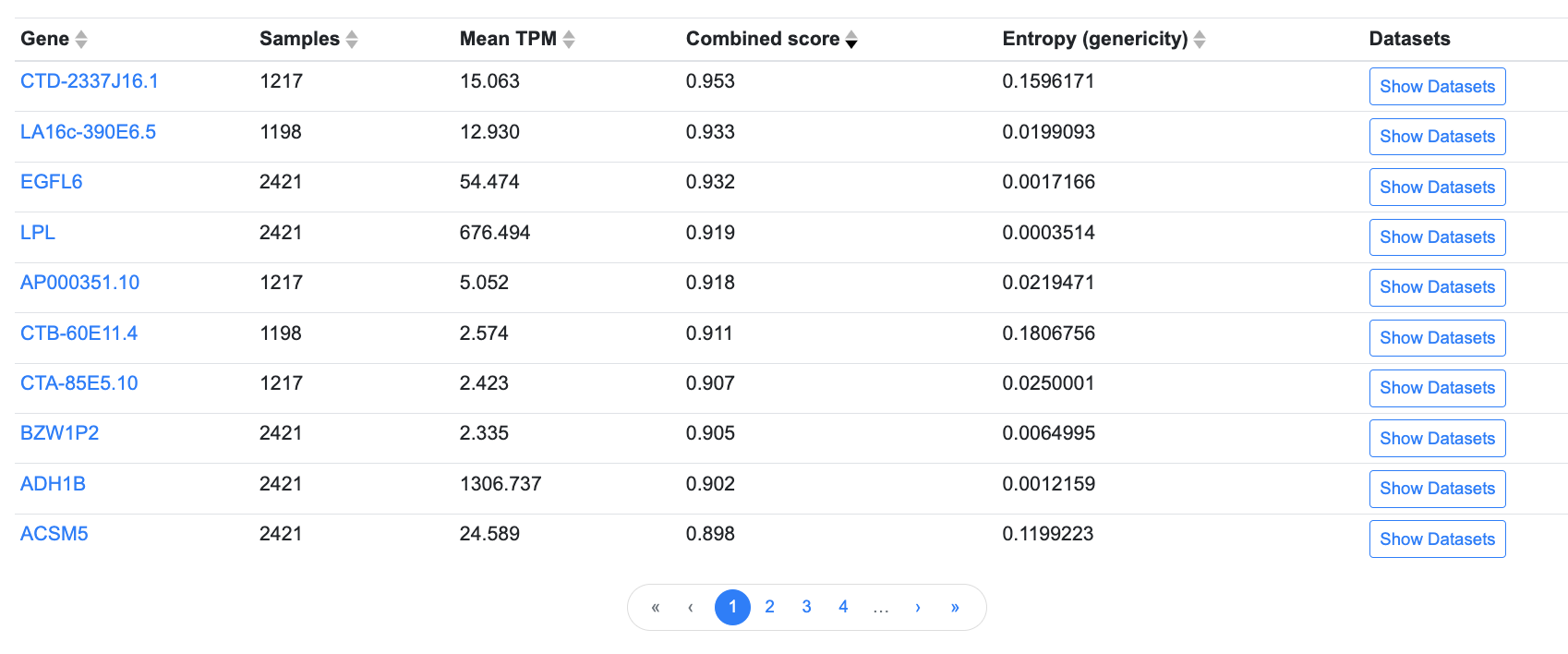
In the table, click on any column header to sort by that column. Use the navigation bar below the table to page through the results.
Click the Show Datasets button to see details about the individual datasets grouped within each tissue category, including their specific tissue or cell type (the Biosample), transcripts per million (TPM) values, and sample sizes. Links in the Dataset column lead to the Common Metabolic Diseases Genome Atlas, where you can see the provenance of the data and view the original datasets. Some datasets represent gene expression in disease states; for those datasets, the disease state is noted in the Disease column.
Credible Sets to Cell Type section
Results from the Credible Sets to Cell Type (CS2CT) method, developed by the Knowledge Portal team at the Broad Institute, can provide clues about the tissues that mediate the effects of causal variants. The method starts with credible sets–both curated from the literature and calculated from meta-analyzed bottom-line associations. The genomic locations of the credible sets are compared with the locations of tissue-specific epigenomic annotations to calculate their overlap. Each credible set is assigned an overlap probability, indicating the likelihood that the causal variant is located within the annotated region, and a “genericity” value that is a measure of the tissue specificity of the overlap.
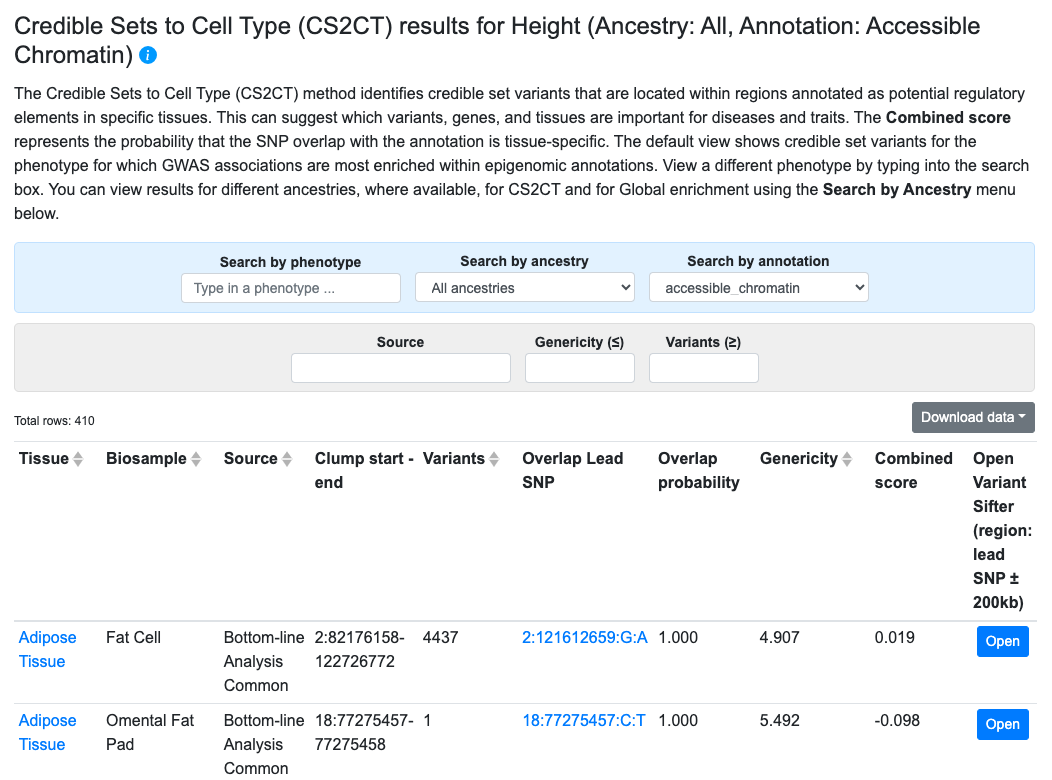
CS2CT results are displayed in a table, as shown above. The results shown by default are those for the credible set and annotation with the highest combined score for this tissue. Metrics for the overlaps are expressed as:
- Overlap probability (higher value = greater probability of overlap)
- Genericity (lower value = greater tissue specificity of the overlap)
- Combined score (higher value = greater probability that the SNP overlaps the annotation in only one or a small number of tissues)
In the table, click the Open button to navigate to the Variant Sifter in the region of a credible set.
Filters above the table allow you to view results for different annotation types, credible set sources, or tissues, and to set a minimum threshold for number of variants in the credible set and a maximum threshold for genericity.
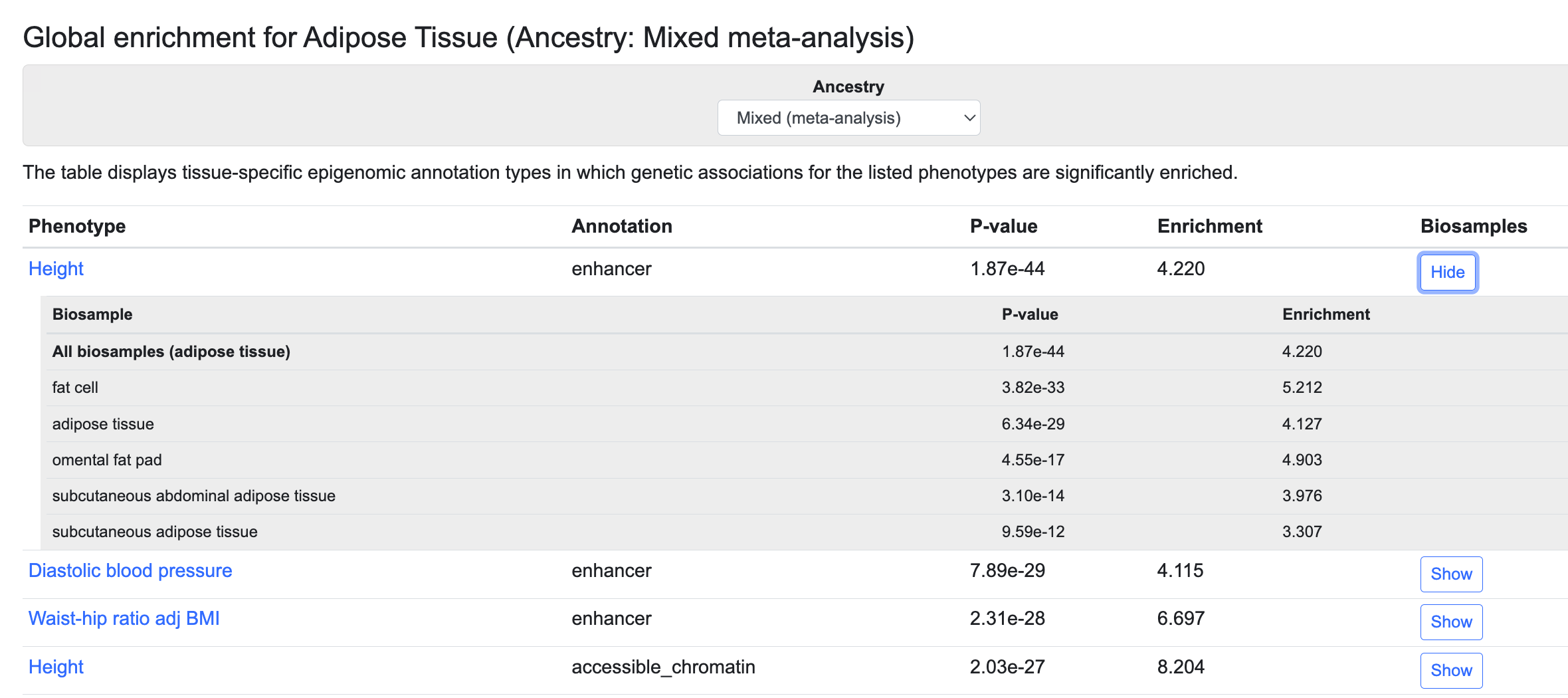
Global enrichment section
This table lists the annotation types that are enriched for genetic associations with the listed phenotypes in the tissue whose page you are viewing, as determined using stratified LD score regression:
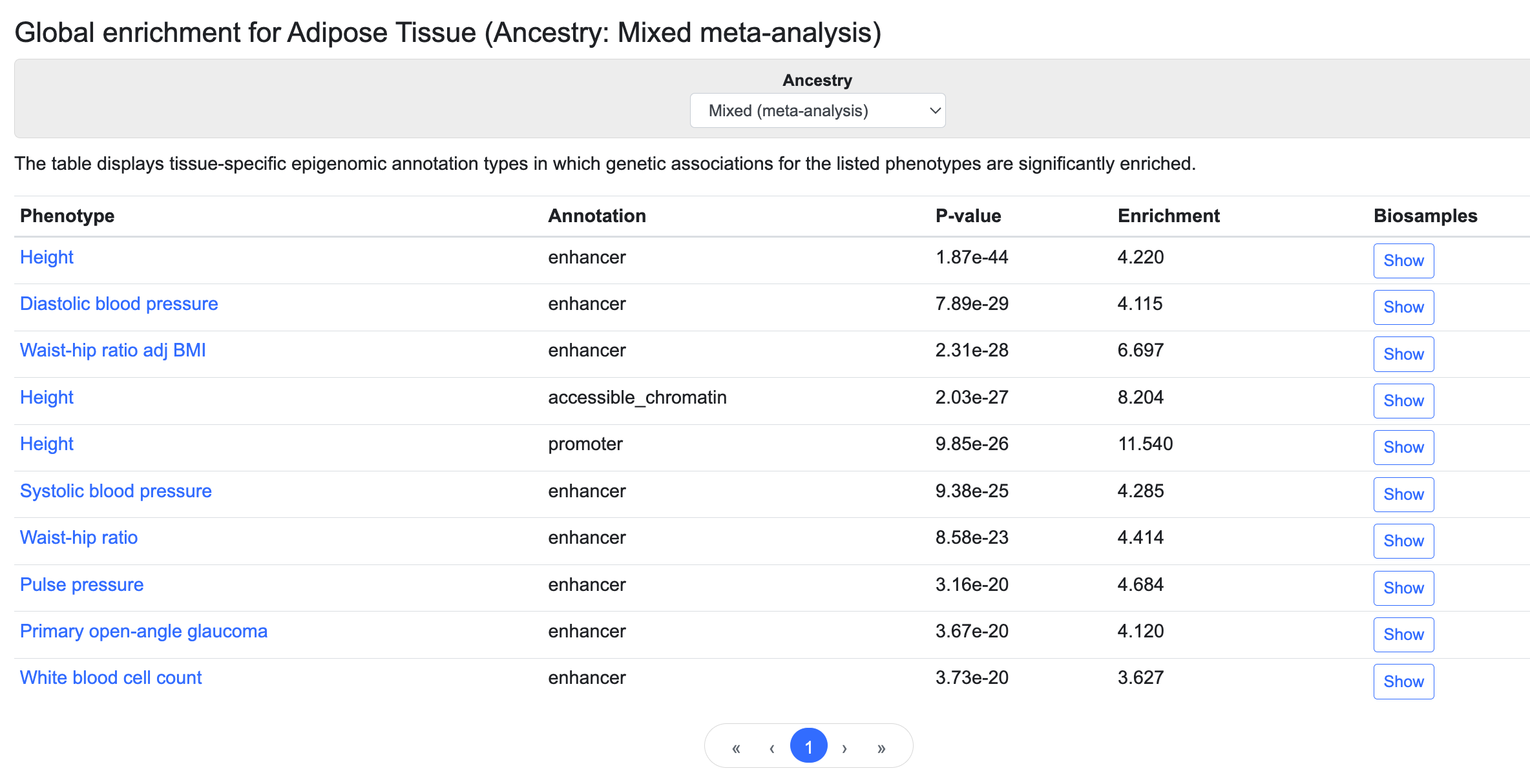
These enrichments can suggest which trait or disease and which regulatory element are most relevant for a tissue. For example, in the table above we see that genetic associations for height are significantly enriched in adipose tissue enhancers and promoters and in regions of open chromatin. The genetic associations are generated by bottom-line integrative analysis, and annotations are derived from the Common Metabolic Diseases Genome Atlas (CMDGA). Global enrichments are available for this set of tissues.
These results are available from a trans-ancestry analysis and from single-ancestry analyses. Use the menu above the table to select an ancestry:
Click the Show button in the Biosamples column to see enrichment values in specific tissue or cell types within the broad tissue category whose page you are viewing.