What can custom computation of gene-level association statistics, based on whole-exome and whole-genome sequencing, reveal about a gene of interest? This worflow illustrates scientific questions that you can explore using the Genetic Association Interactive Tool (GAIT).
The figure and video below illustrate the overall GAIT workflow:
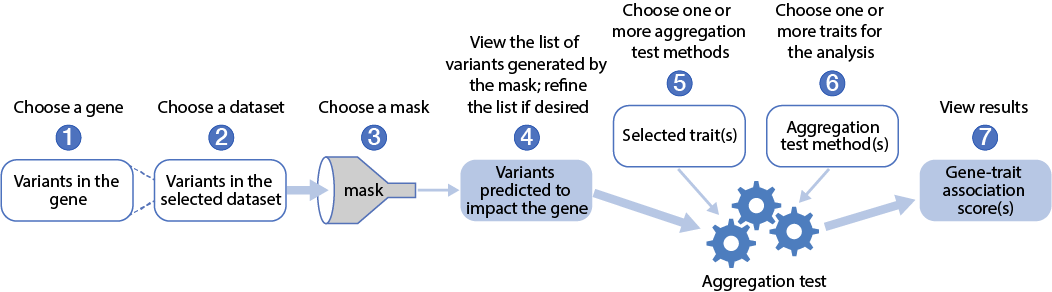
The data sources and methods used in GAIT and how to use the interface are described in detail in the GAIT Guide. Note that the GAIT interface is currently only available in the Knowledge Portals that include diabetes and cardiometabolic traits.
One potential use of GAIT is to use it to test whether a specific domain of a protein may be responsible for its role in a disease or trait. As an example, we'll look at the HNF1A gene, which is known to have a role in type 2 diabetes (T2D).
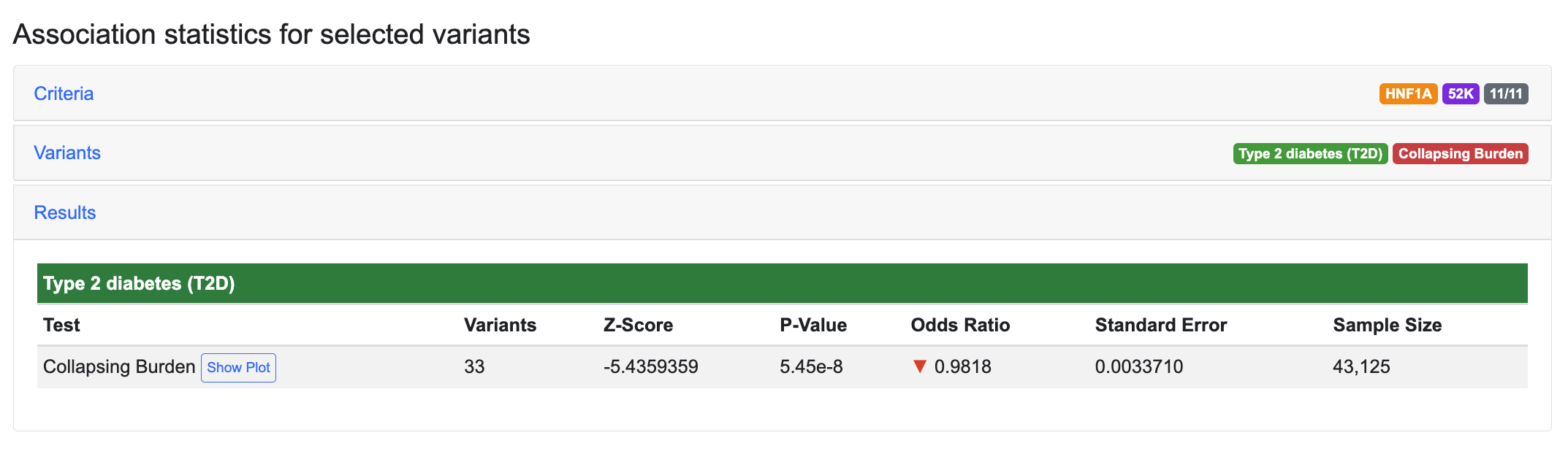
Using GAIT to analyze type 2 diabetes (T2D) association of HNF1A with the 11/11 mask and the 52K dataset, we retrieve 60 variants. Running the collapsing burden test produces a p-value of 5.45e-8 for association of HNF1A with T2D (Navigate to the GAIT interface with these parameters):
The transactivation domain of the HNF1A protein (residues 287 to the C terminus) is known to be important for its function. If we repeat the analysis, selecting only the following variants and de-selecting the other variants retrieved by the mask:
- All nonsense and frameshift variants in the transactivation domain
- Two mutations known to cause maturity-onset diabetes of the young (MODY) that have been experimentally shown to severely decrease HNF1A activity: P112L and P447L (Najmi et al, 2017)
...the significance of the T2D association is increased, with the p-value decreasing to 3.41e-8:
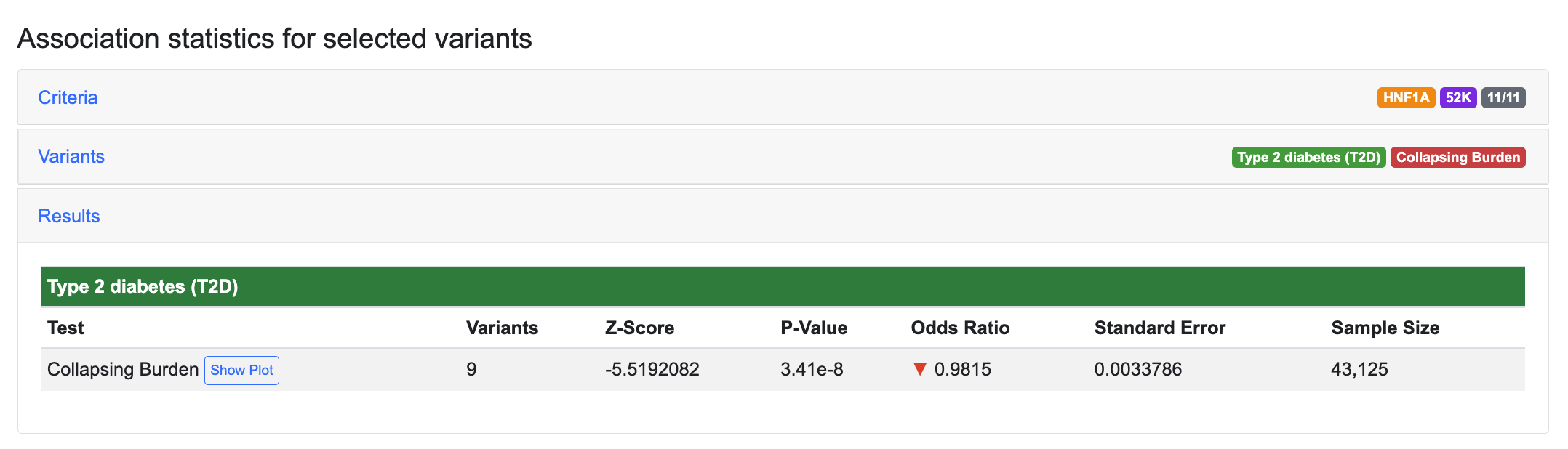
This illustrates how selecting a set of functionally important variants can produce more meaningful results with GAIT.
Another use of GAIT is to refine association signals in order to determine which variants are most important to an association. As an example, we'll do a custom association test for the MC4R gene. We'll use the AMP T2D-GENES exome sequence datasets as input ("52K" in the Datasets menu) and the 5/5 mask to filter variants.
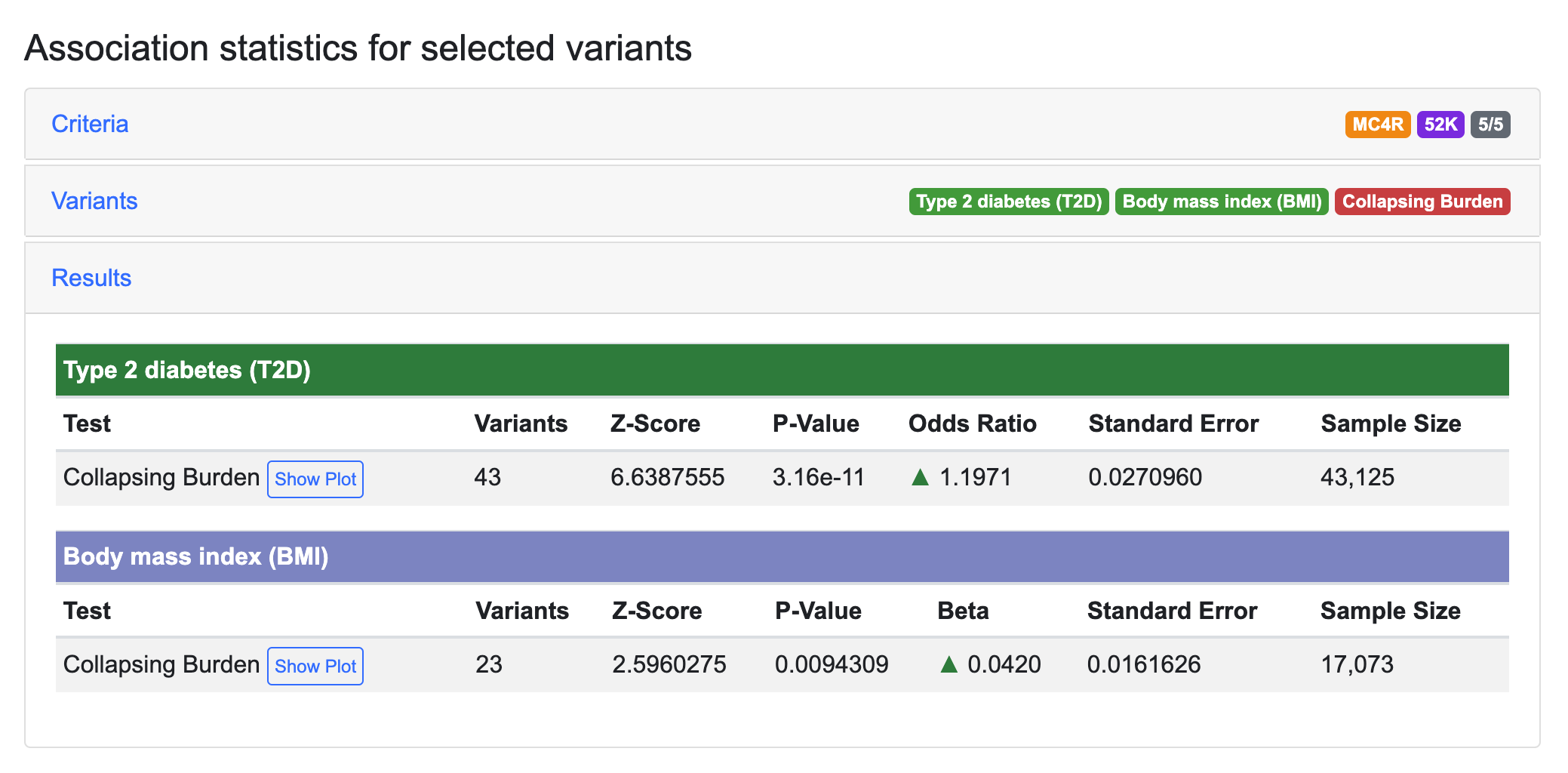
Analyzing the same exome sequence data and using a slightly different methodology, Flannick et al., 2019 published similar results for gene-level associations of MC4R with type 2 diabetes and BMI, and reported that much of this association signal is due to one variant, 18:58038777:A:T (rs79783591), which causes an Ile269Asn missense change in the MC4R protein.
GAIT can be used to conduct the gene-level association analysis while excluding this variant, by de-selecting it in the variant table:
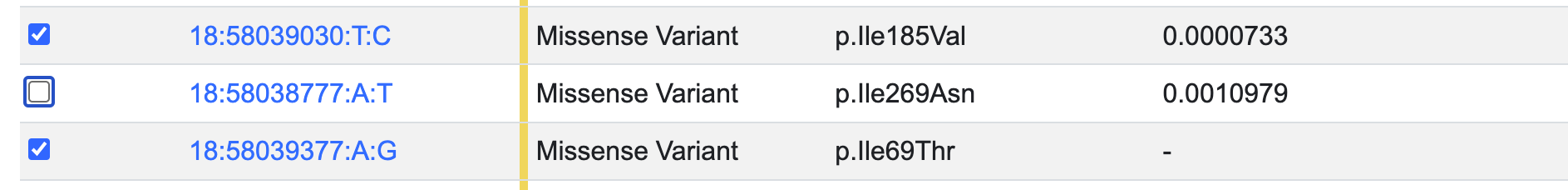
After re-running the analysis, we see that GAIT replicates the published result: for the variants in this mask, nearly all of the BMI signal and most of the T2D signal were due to rs79783591.
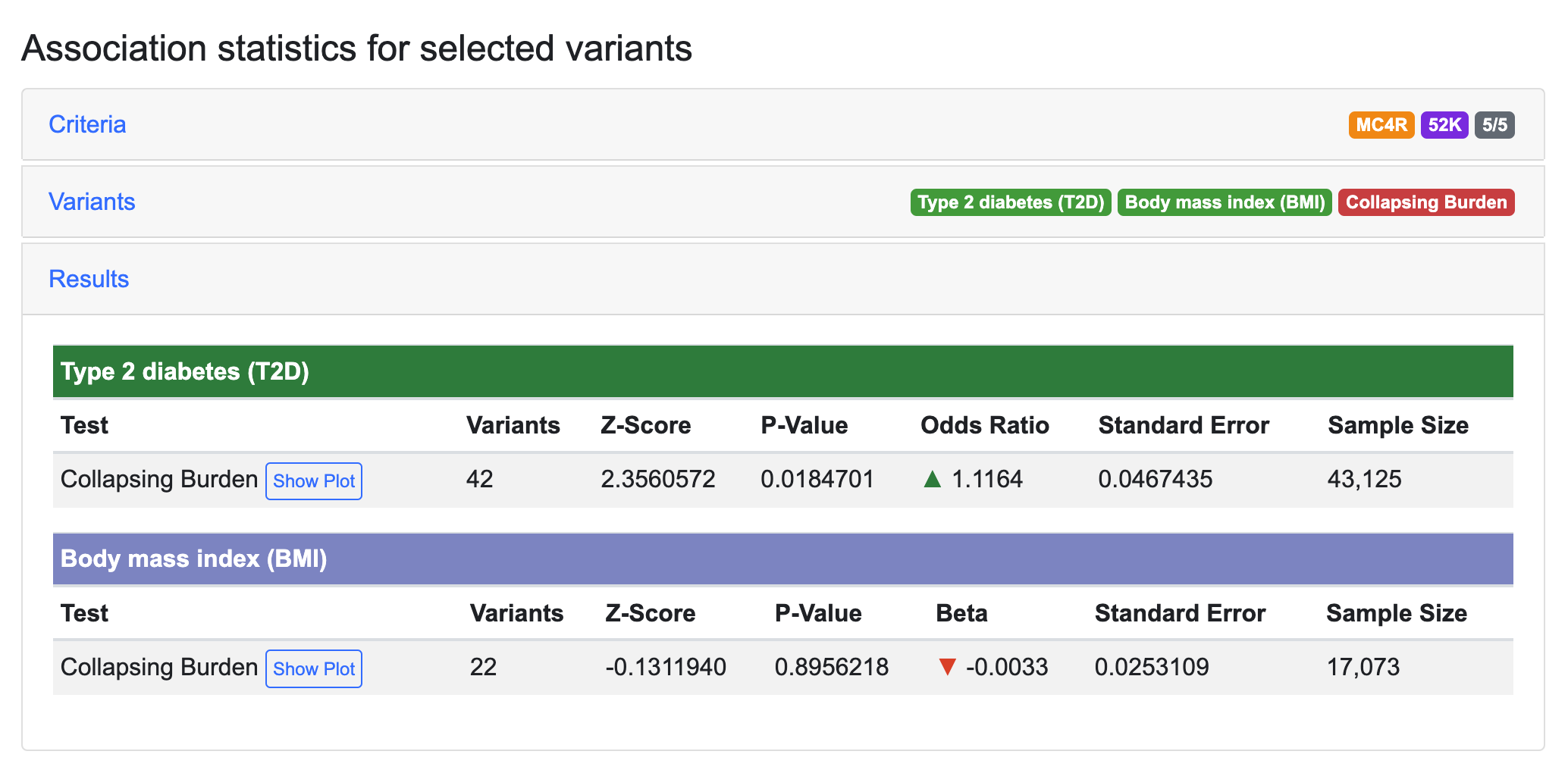
This illustrates how GAIT can be used to dissect the contribution of specific rare variants to aggregate association signals for a gene.
