Predicted effector lists can give you an overview of the genetic architecture of a disease or trait.
What is a predicted effector list?
Where can I find effector lists?
How do I navigate individual effector list pages?
View a tutorial video on predicted effector lists:
What is a predicted effector list?
The ultimate goal of genetic association studies is to discover which genes and pathways have direct roles in risk of a disease or trait. Many disease-focused studies now include predictions of the most likely effector genes for that disease. The methods used to generate these predictions vary widely. Most start with a GWAS locus and then integrate genetic evidence (coding variants; related monogenic diseases) with regulatory evidence (molecular QTLs; tissue-specific chromatin conformation) and other biological evidence (model organism phenotypes; drug interactions; results of methods such as DEPICT and PoPS; results from the literature) to predict which gene at the locus is likely to have a direct impact on disease risk.
These lists are valuable because they represent the current thinking of experts in each disease community, but they are often not open-access, or they are difficult to find within the large number of supplementary files that accompany most publications. We are endeavoring to make these lists accessible and interactive within the Knowledge Portals (KPs) by collecting all available lists for traits and diseases relevant to the KPs, representing them in interactive tables (see below), and connecting them to genes and phenotypes in the Knowledge Portals.
Currently, in the first phase of this project, we are collecting all available lists for traits and diseases relevant to the KPs, representing them in interactive tables, and making them accessible on Gene pages and in the Gene Sifter tool. In the future, we will improve the accessibility of the lists by developing ways to compare them and to search across lists for genes of interest.
Have you generated a list of predicted effector genes for a common disease or related trait? Please contact us to have the list displayed in the Knowledge Portals.
Where can I find effector lists?
The examples in this tutorial show the Common Metabolic Diseases Knowledge Portal (CMDKP), but the workflow illustrated is applicable to most other KPs.
To view lists of predicted effector genes, click the Predicted Effector Genes link in the upper KP Labs menu:
On the resulting page, you can browse all available lists, or you can narrow down which lists to view by entering filtering criteria:
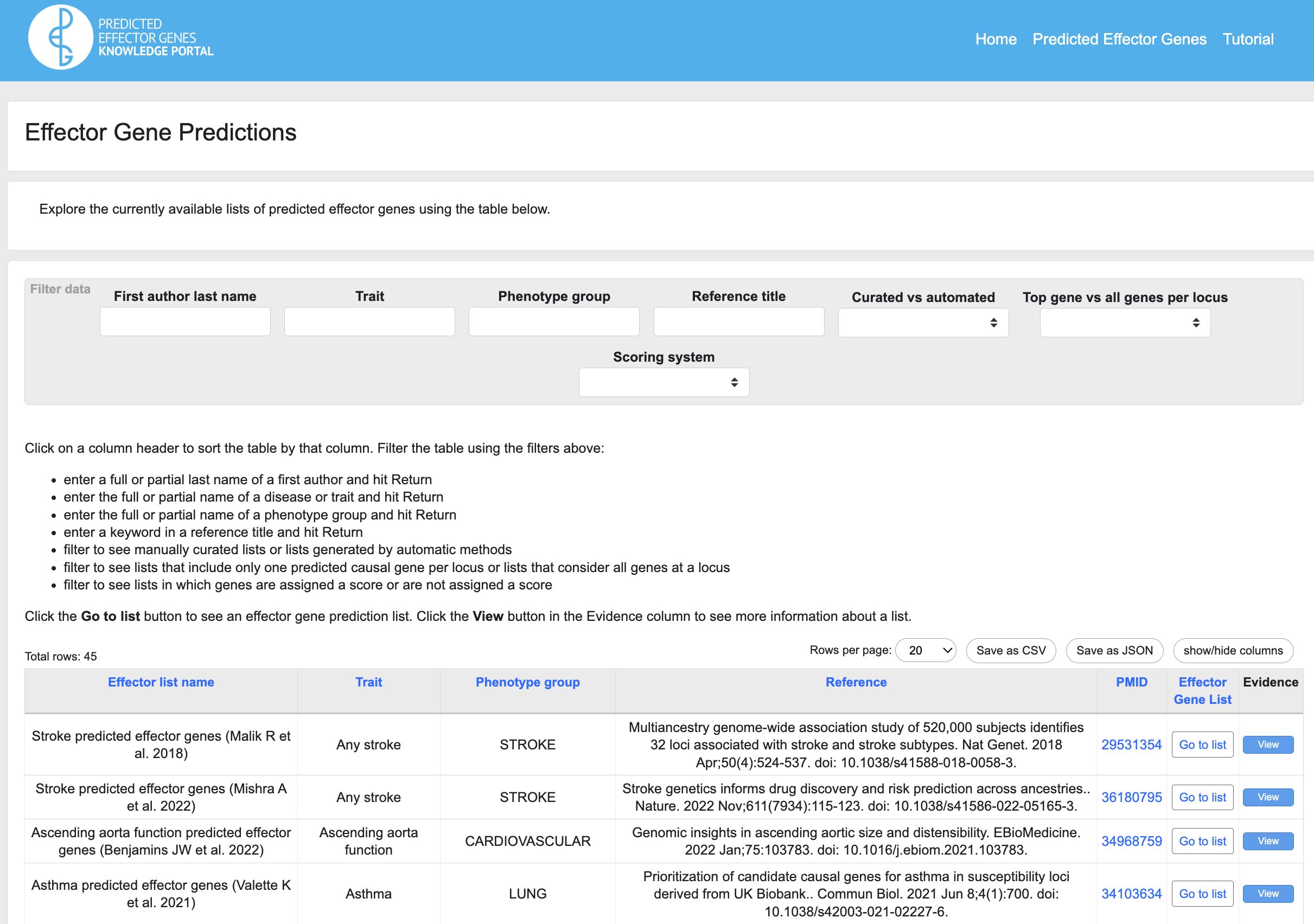
Use the filters to search and navigate the lists:
- The First author last name filter searches the Effector list name column, which includes the last name of the study's first author. Enter a full or partial last name and press Return to view the rows that include your search criterion.
- The Trait filter searches the Trait column, indicating the trait or disease for which effector genes were predicted. Enter a full or partial trait name and press Return to view the rows that include your search criterion.
- The Phenotype group filter searches the Phenotype group column, which indicates the general categorization of the trait or disease for which effector genes were predicted. Enter a full or partial phenotype group name and press Return to view the rows that include your search criterion.
- The Reference title filter lets you search for full or partial keywords in reference titles.
- The Curated vs Automated filter lets you select lists that were generated by automatic methods from those that incorporate multiple independent evidence types and expert judgement.
- Some lists include only the top predicted causal gene at each GWAS locus, while others include all genes at each locus. The Top gene vs all genes per locus filter separates the two types of list.
- Some lists score genes by the weight of evidence supporting the prediction that they are effector genes. The Scoring system filter lets you see which lists use a scoring system.
Multiple filters may be combined to narrow the list.
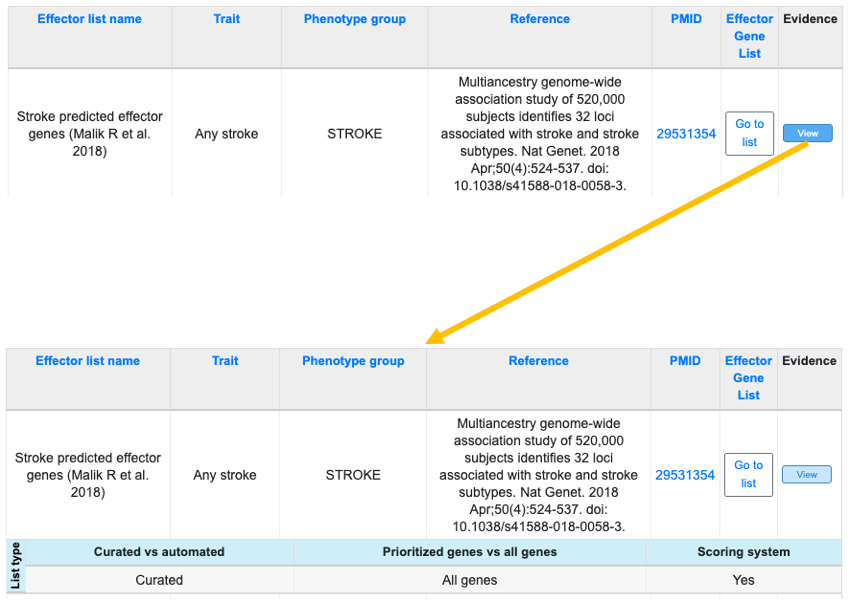
In the table of predicted effector lists, click the Go to list button to navigate to a list, or click the View button in the Evidence column to see whether a list was generated by manual curation or automated methods, whether it includes top genes only or all genes at GWAS loci, and whether a scoring system was used.
How do I navigate individual effector list pages?
The effector lists differ in their formats because the supporting evidence types differ between lists, but there are commonalities. We'll illustrate them using a list of predicted effector genes for estimated glomerular filtration rate.
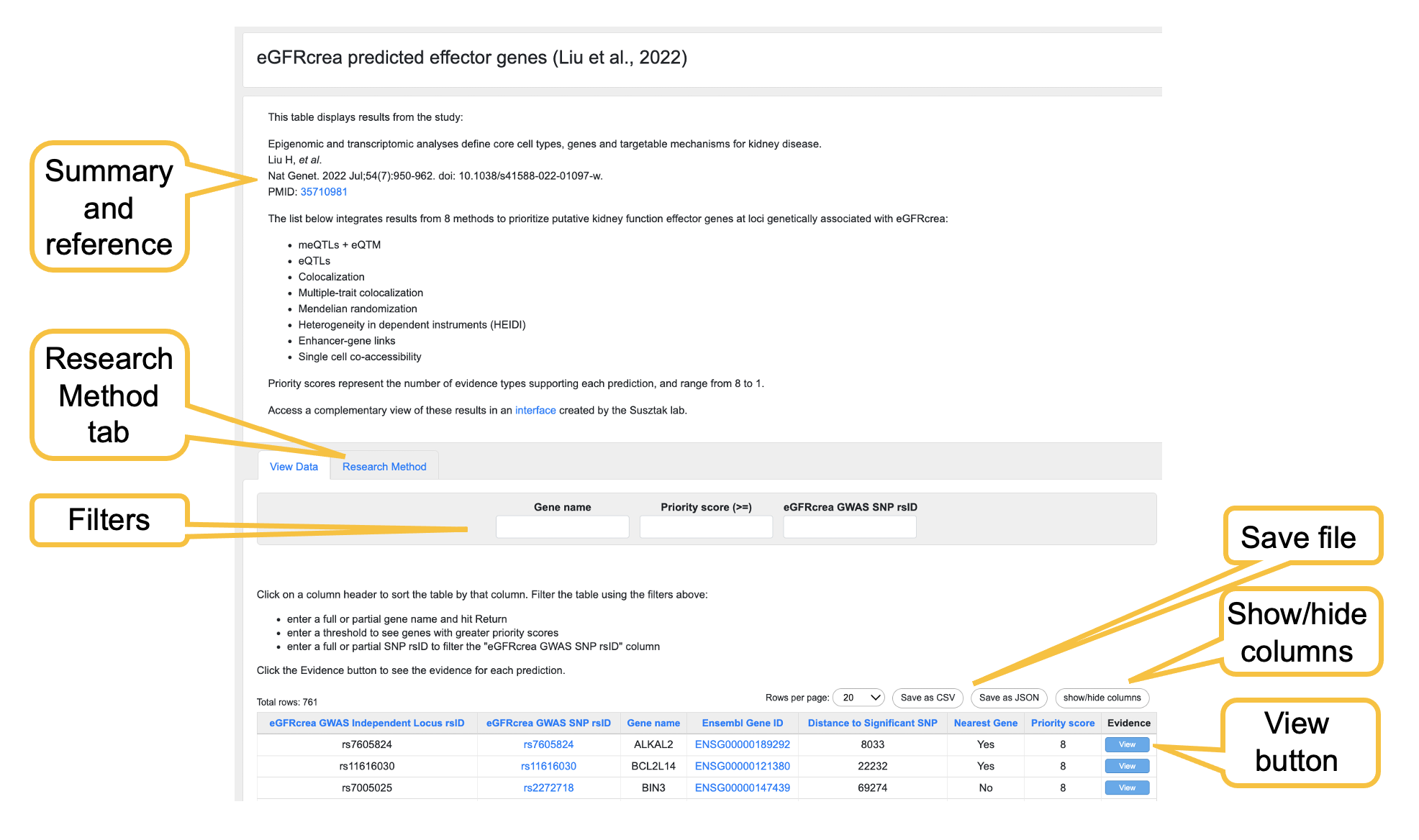
As illustrated in the figure above,
- The top of the page shows the reference in which the list was published (if applicable), and briefly summarizes the prediction method.
- Some lists include a more detailed description of the method and supporting data that can be accessed using the Research Method tab.
- In the table, click on a column header to sort the table by that column. Mousing over some column headers will display a tooltip with more explanation about the data in that column.
- Custom filters allow you to explore the results in the table. There are 3 types of filters:
- Text search: enter a text string and press the Return key, to display only rows that include that string. For example, entering "ra" into the Gene name filter would display only rows in which a gene name in the Gene name column includes "ra".
- Search greater than/less than: allows you to set a threshold for numerical values. In the example list shown, entering "6" into the Priority score (>=) filter would display rows with priority scores 6, 7, and 8.
- Drop-down menus: allow you to make a selection from a menu to display the rows that include that value or text string.
- You may download the file in .csv or JSON formats.
- Clicking the Show/hide columns button displays a list of the columns displayed by default and allows you to customize it.
- The View button expands the row to display one or more nested rows beneath it that include supporting evidence for the prediction.
