The Variant Sifter allows you to explore genetic associations, credible sets, and multiple types of epigenomic annotations to prioritize potentially causal variants and genes in a locus. In this example, we'll explore potential causal variants for type 1 diabetes (T1D) near the BACH2 gene. We'll do this in the Type 1 Diabetes Knowledge Portal (T1DKP), but this workflow is applicable to most other Knowledge Portals.
You can navigate to the Variant Sifter interface directly from the upper Tools menu and enter your phenotype and gene or region of interest, or you can start from a Region page that displays the region, phenotype, and ancestry that you have selected.
We start by entering BACH2 into the T1DKP home page search box to navigate to a Region page around that gene, and then scroll down to the Genomic Region Miner (GEM) module. By default, GEM displays associations for the most significantly associated phenotype in the region--in this case, Eosinophil count. We'll add the Type 1 diabetes phenotype using the Add phenotypes menu above the LocusZoom plot:
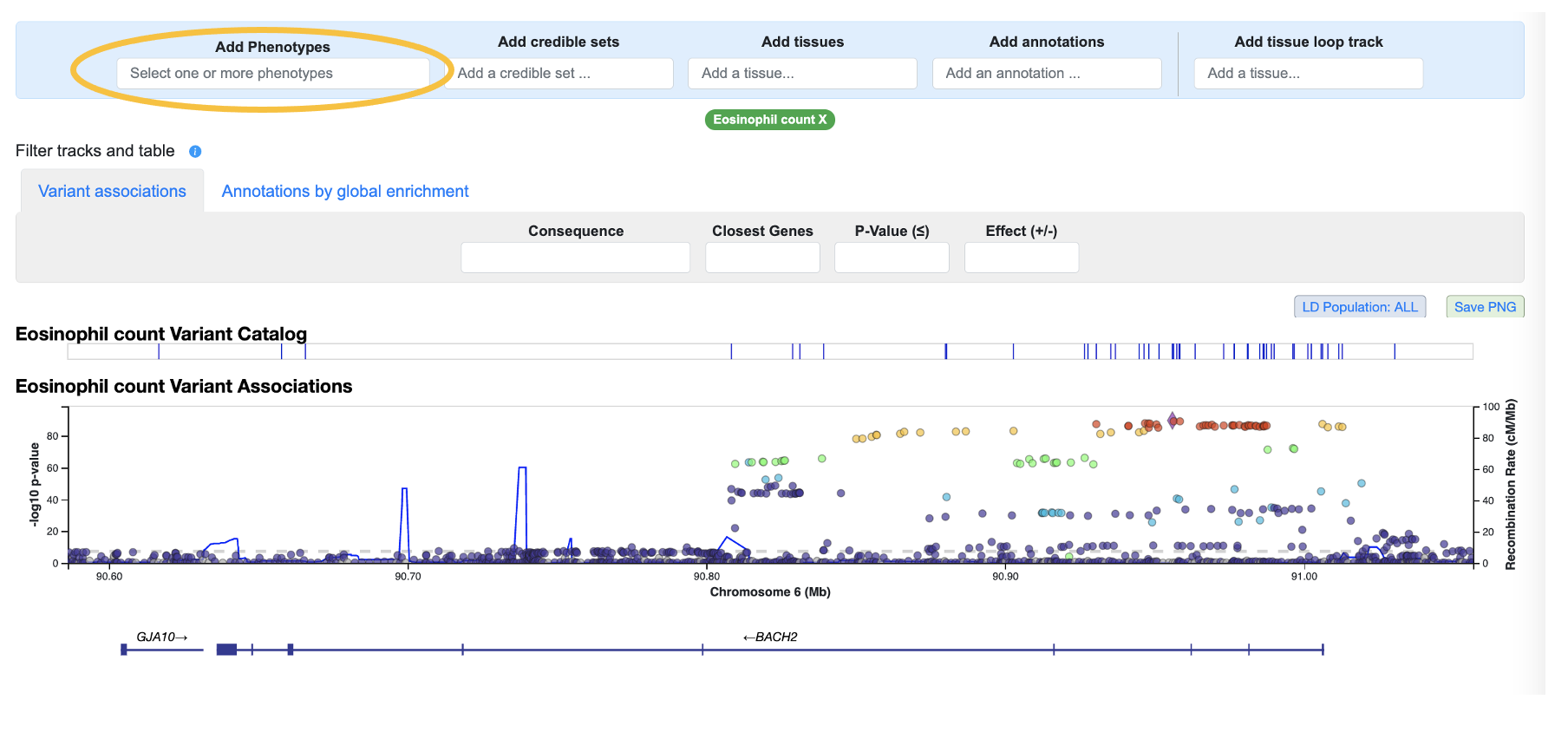
We can then remove the Eosinophil count phenotype by x-ing out that phenotype in the interface:
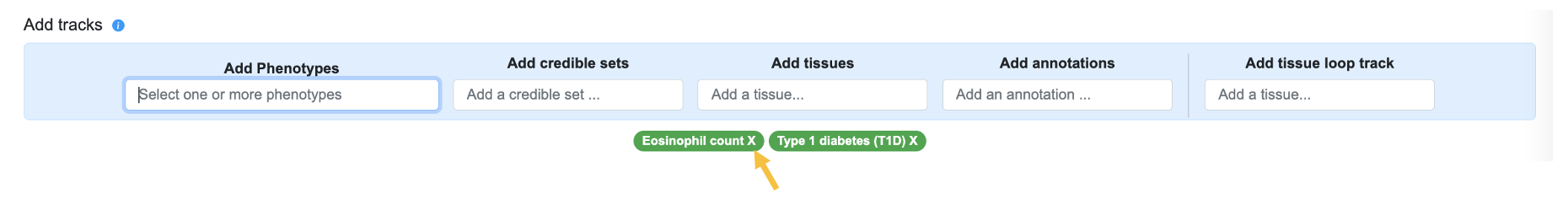
Note that the Variant Sifter can display associations for multiple phenotypes, and also for specific ancestries (if you select an ancestry in the Set page level parameters box at the top of the page), but we'll keep it simple for this example.
Next, click the Prioritize variants in this region link below the LocusZoom plot.
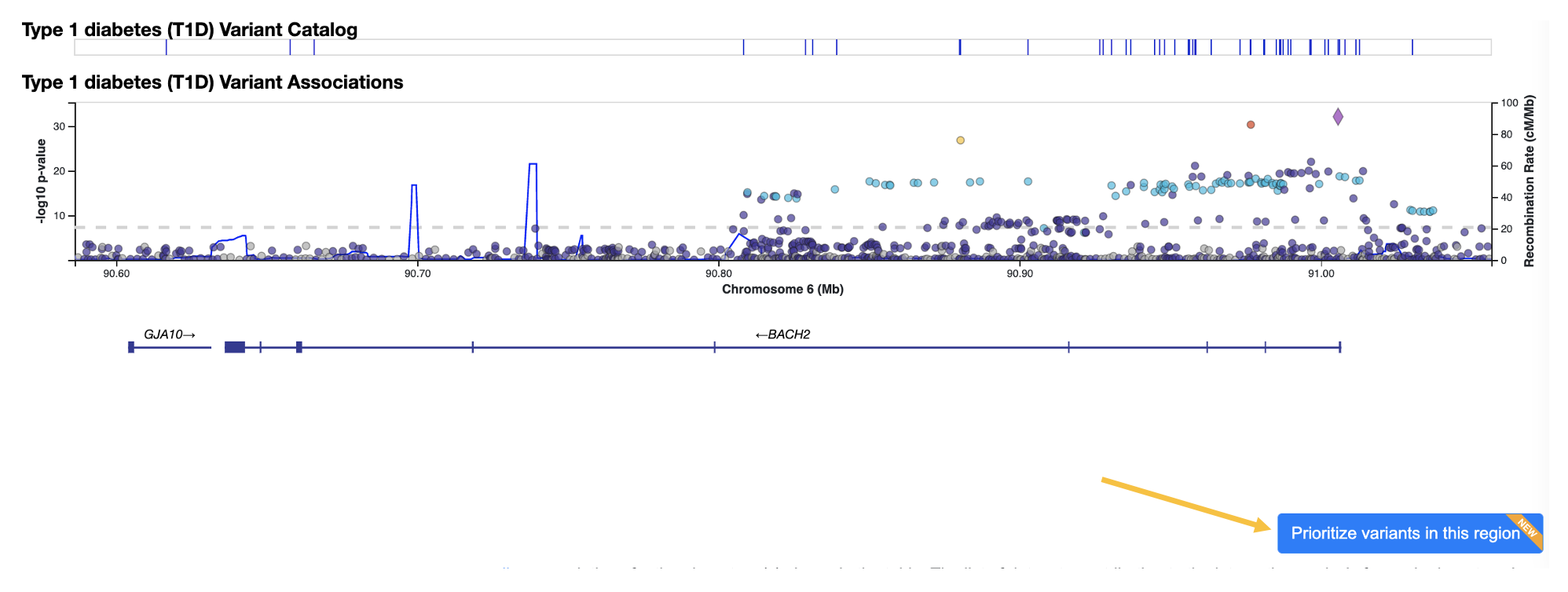
This takes you to the Variant Sifter page. The initial view shows T1D associations across the BACH2 region. All 10,463 variants with p < 1.0 are listed in the table below.
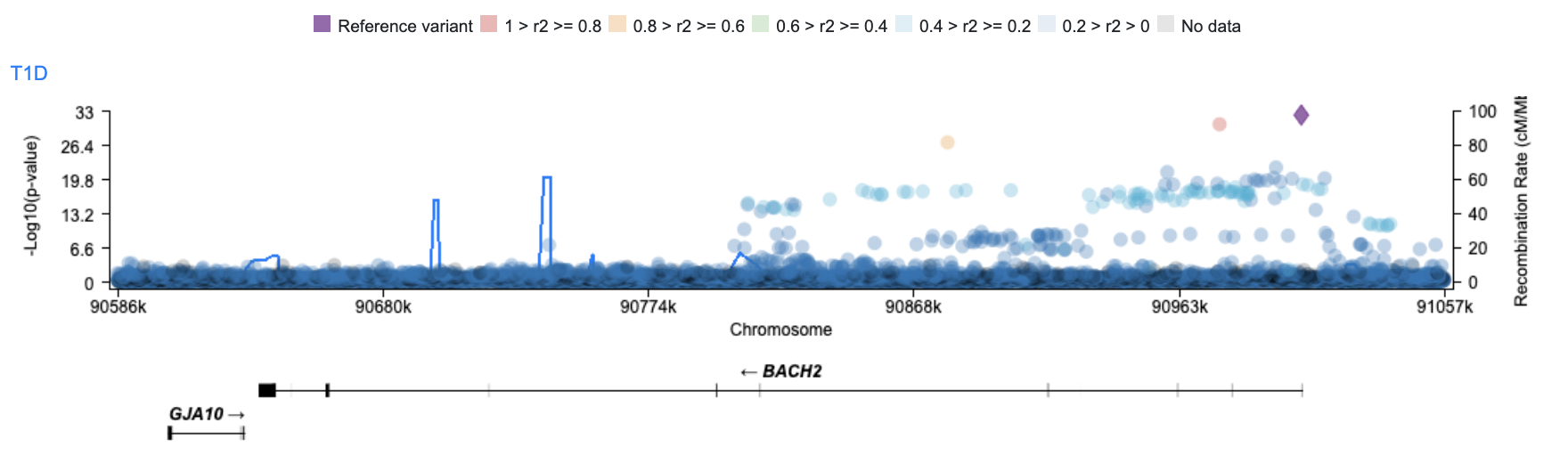
We suspect that some of the most significantly associated variants may be important. To keep track of them through the rest of the workflow, we can mark them with a star. Click on a variant of interest in the association plot to open a tooltip, then click the star to mark a variant.
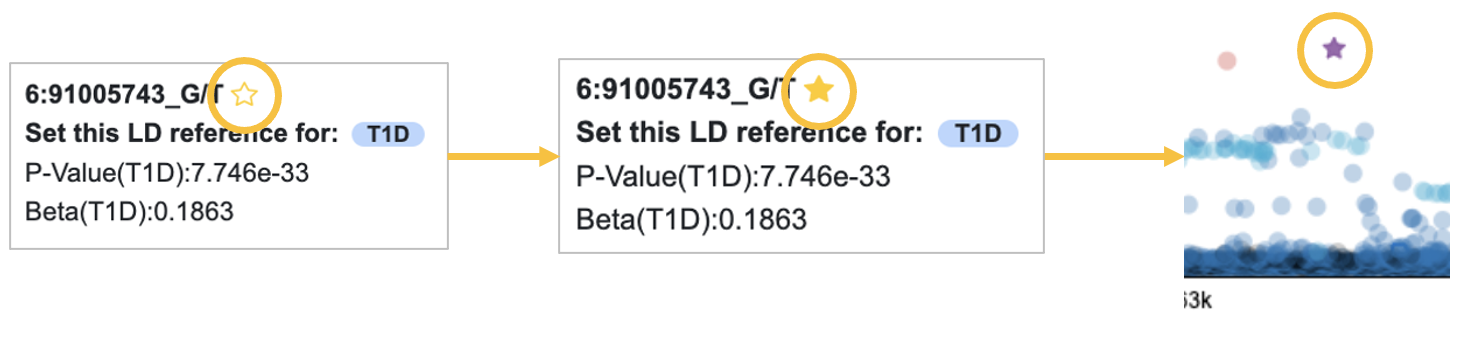
We'll star three of the top variants in this region.
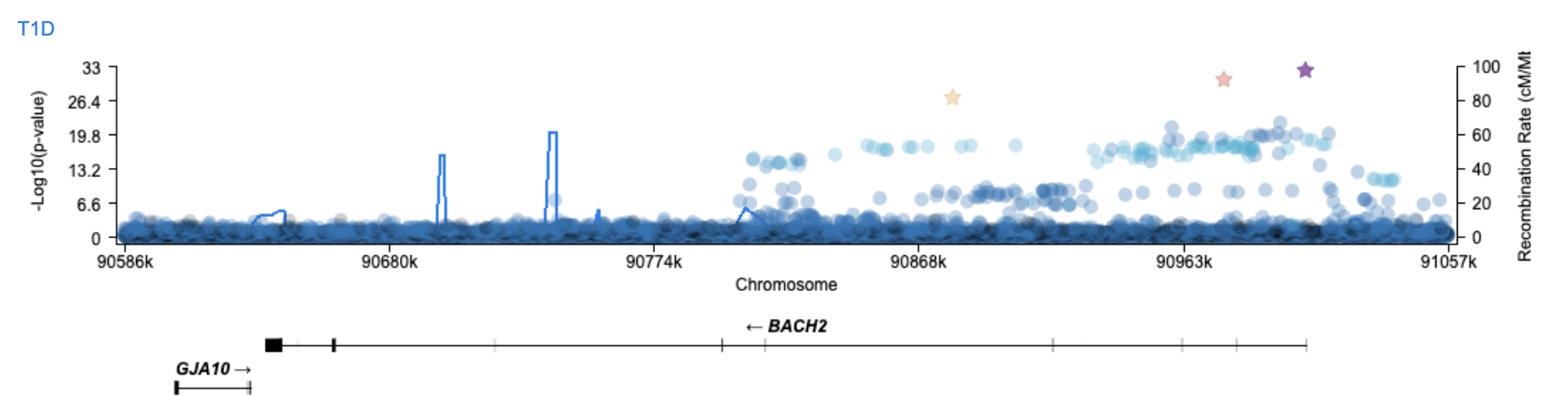
Genetic associations alone can't tell us which among the 10,463 variants in this region is the causal variant, so the Variant Sifter allows you to integrate other data types that can provide supporting evidence.
Credible sets are sets of variants, defined by investigators, that are predicted to include the causal variant for an association signal. If credible sets are available for the region and phenotype you've chosen, you can opt to display these below the LocusZoom plot. In our example, there are two credible sets available for T1D, both from Chiou et al., 2021.
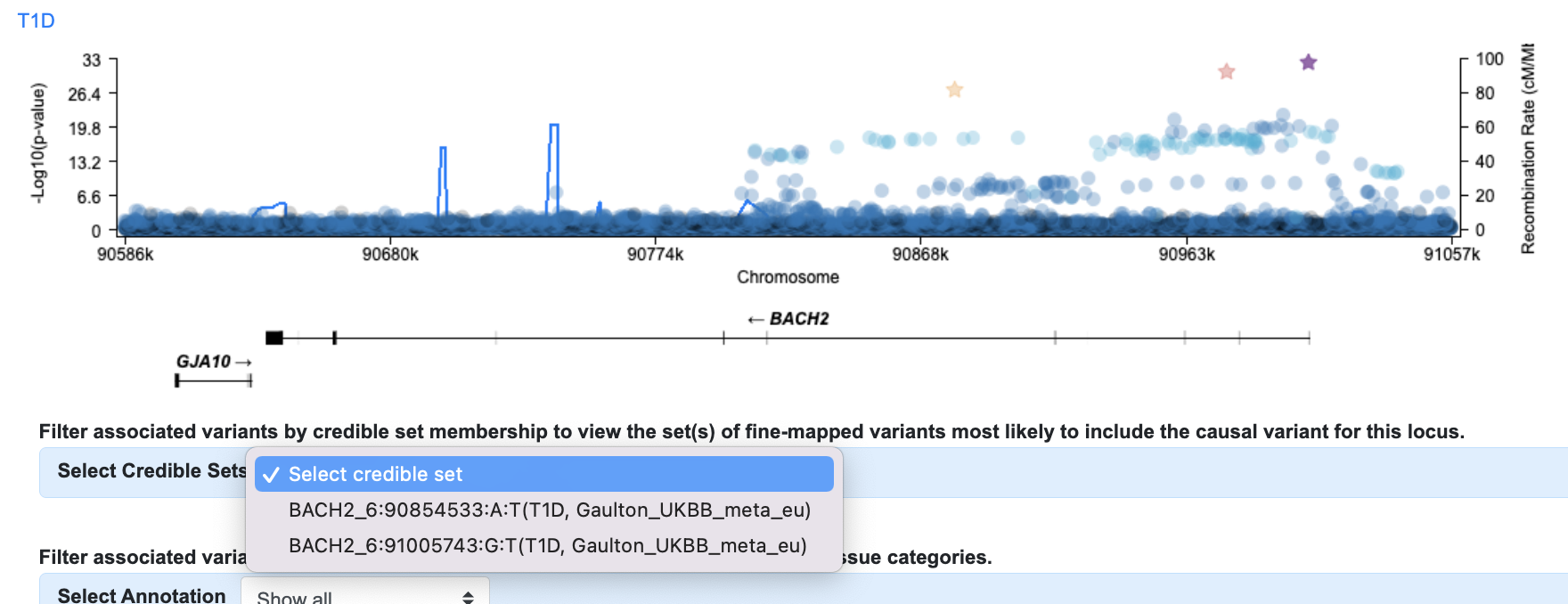
If we select both sets, they are both displayed, with different colors:

The variant table at the bottom of the page is now filtered down to the 139 variants that belong to either of these credible sets.
Now we'll add a layer of genomic annotations. These can suggest the regulatory potential of a region, so if a variant is located within an annotated region that can suggest hypotheses about the mechanism by which it affects disease risk.
The Global Enrichment plot can suggest which tissues may be most relevant for the phenotype you're viewing. The values in the plot were calculated from bottom-line ancestry-specific genetic associations by the GREGOR (Genomic Regulatory Elements and Gwas Overlap algoRithm; Schmidt et al., 2015). Significant enrichment of genetic association signals for a disease in annotated regions of a particular type in a specific tissue suggests that the tissue may be relevant for the disease.
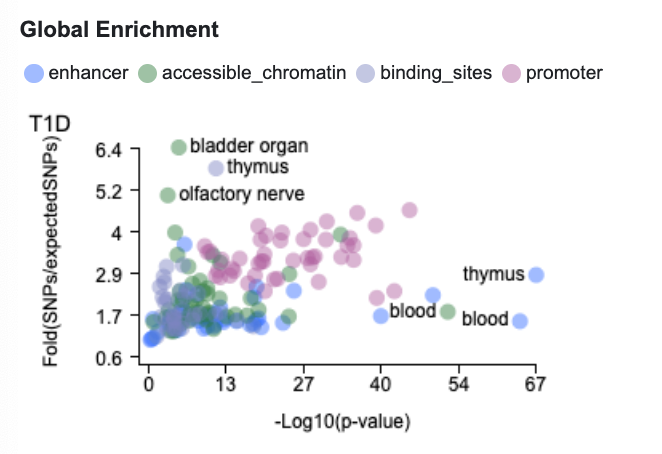
In the context of the Variant Sifter, the Global Enrichment plot can suggest which annotations and tissues may be most relevant to view for a particular phenotype. In this case, we see that T1D genetic associations are most significantly enriched in regions annotated as enhancers in the broad tissue categories thymus and blood, so let's look at the pattern of enhancer annotations across the region. Note that the plot represents genome-wide results rather than enrichment results for this specific region.

Here each track represents the pattern of regions annotated as enhancers in a broad tissue category. The locations of our starred variants are marked by vertical dotted lines. If we click on a track, it becomes highlighted:

...and the annotations in the specific cell and tissue types within that broad tissue category are displayed below:
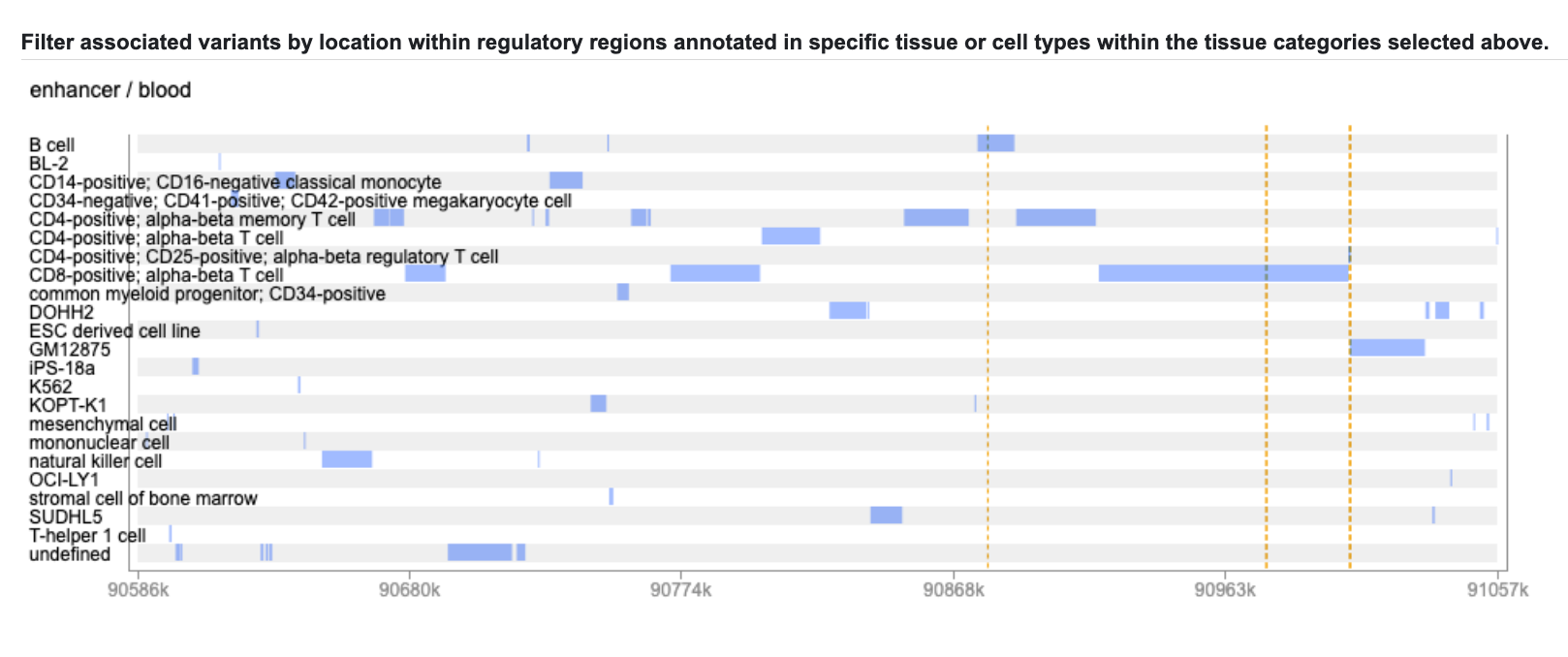
We see that the variants we've highlighted are indeed located within predicted enhancer regions in some immune cell types. If you mouse over the tracks you can see details about each annotation:
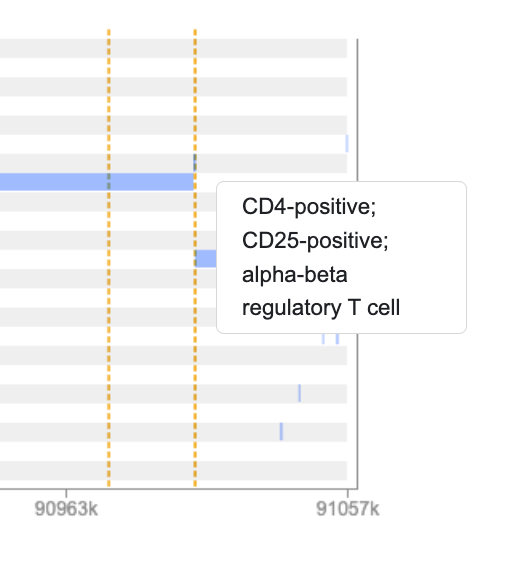
Click on one or more tracks to highlight them:
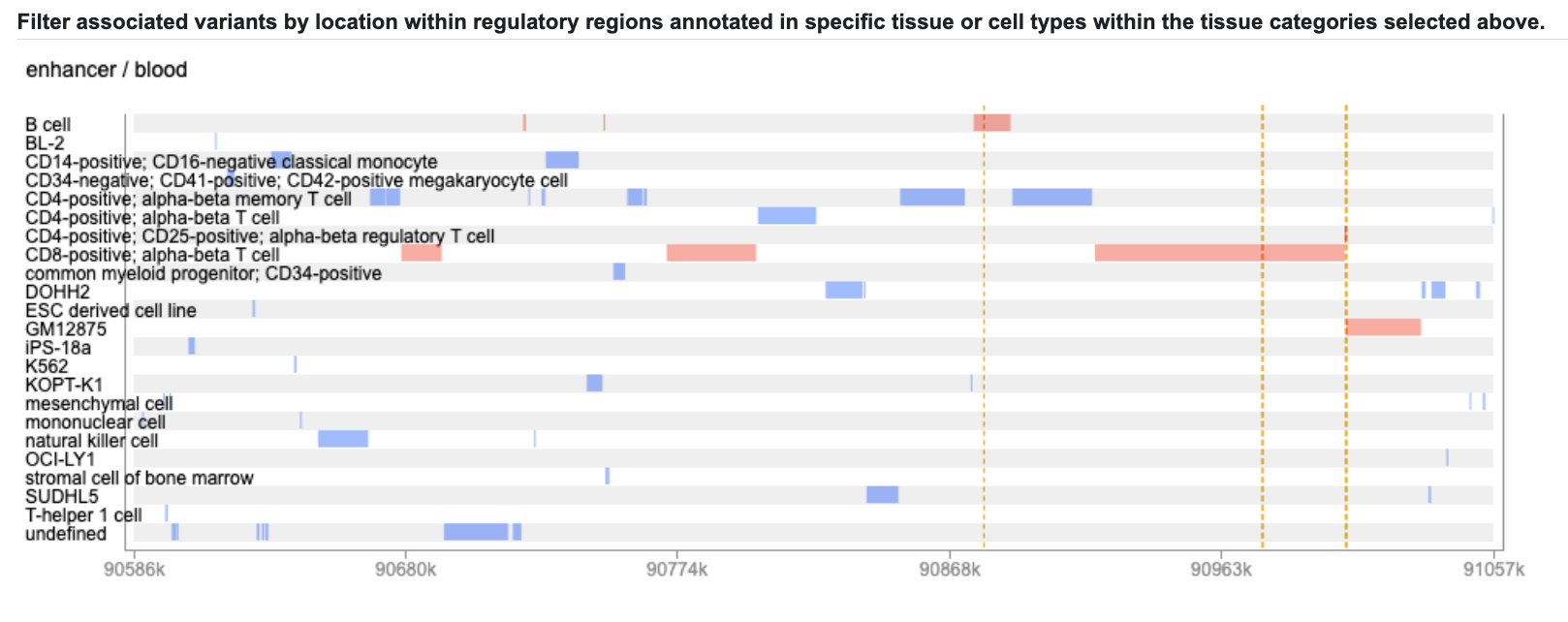
Now the details are displayed in the table below, along with links to view the original datasets in the Common Metabolic Diseases Genome Atlas:
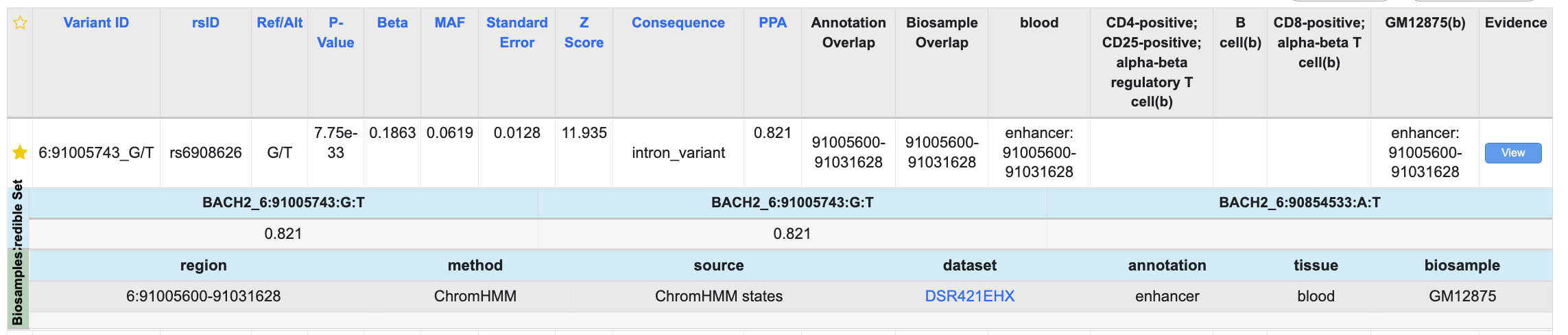
The table now displays only the variants that meet all the criteria you've selected so far: genetically associated with T1D, present in a T1D credible set, and located within a region annotated as an enhancer in the specific blood cell types that are highlighted.
Finally, we can investigate whether any of our variants of interest lie within annotations generated from chromatin conformation assays that link the BACH2 gene to genomic regions. We select "blood" as the tissue in the Target Gene Links filter:

...and now we see the genomic regions that are predicted to be contact the promoters of specific genes:
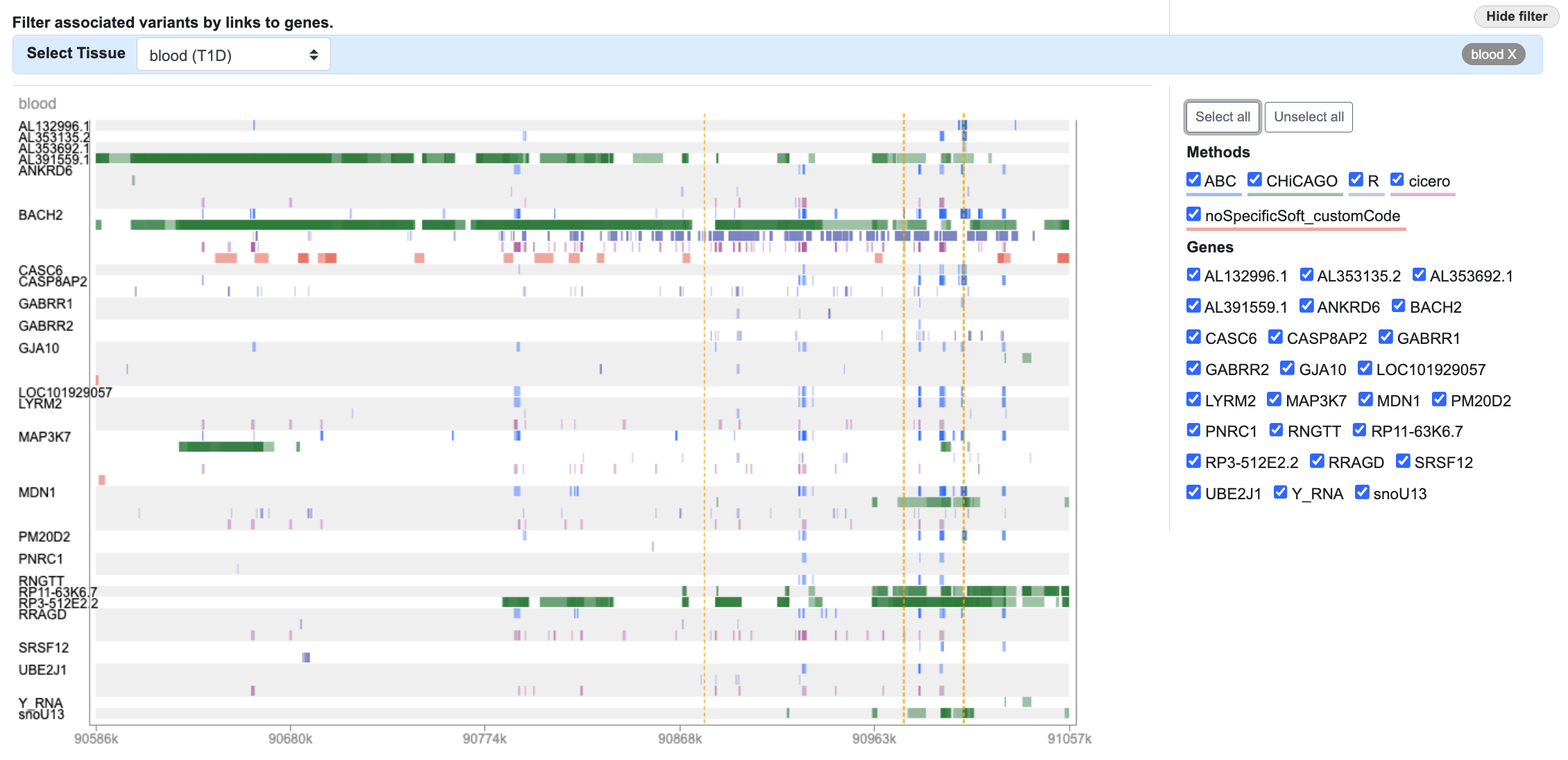
To focus on BACH2, we'll click Unselect all on the right, and then re-select BACH2 and all of the methods:
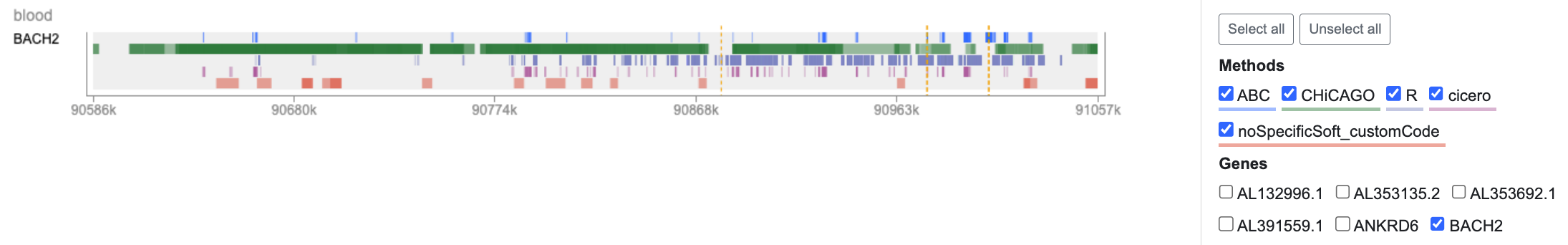
Mousing over the annotated regions, or viewing the table below, shows us that two of our variants of interest are indeed predicted, by several different methods, to be linked to BACH2.
Our results with the Variant Sifter are consistent with the published literature: one of the top two variants seen in our example was found by Robertson, Inshaw, et al., 2021 to impact BACH2 regulation in immune cells.
We hope that you find the Variant Sifter useful. We're continually adding more data and new data types to it, and the interface is under active development. Please contact us if you have questions or suggestions!
