The Knowledge Portal genetic support workflow can help answer the question: "Do human genetic and genomic results provide evidence about the involvement of a gene of interest in a disease or trait?". Based on an analysis of recently published papers that proposed roles in metabolic disease for specific genes, researchers do not often cite evidence from human genetics in support of their experimentally derived hypotheses (Dornbos et al., 2022). This is likely because human genetic results have historically been difficult to access and their interpretation can be challenging for researchers not trained in genetics. The Knowledge Portals aim to reduce these barriers so that all kinds of research can benefit from human genetic findings.
A summary of the workflow:
Get an overview of the workflow in this video:
Step-by-step guide
This guide will use "phenotype" as a generic term to denote a disease or trait. Although the screenshots show the Common Metabolic Diseases Knowledge Portal, this workflow is applicable to most other Knowledge Portals.
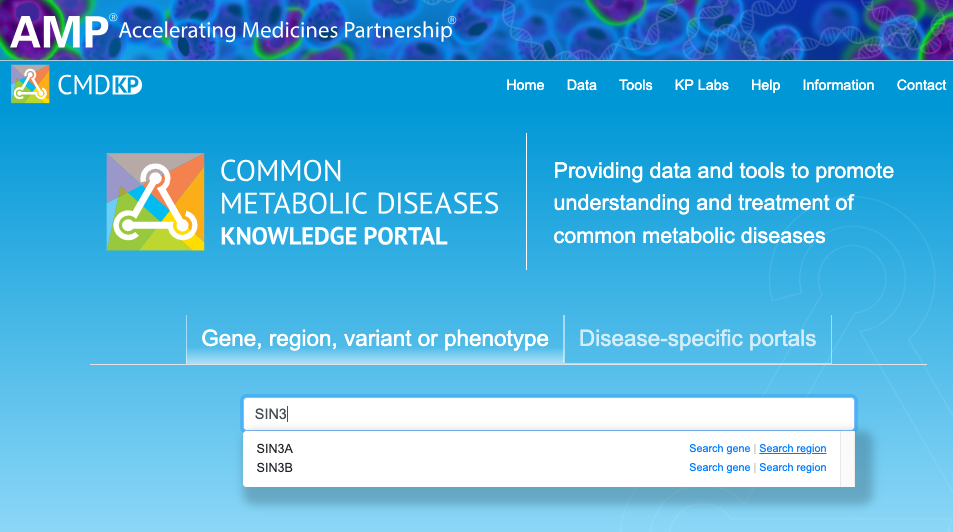
To start: Navigate to the Region page around a gene of interest. Starting on the portal home page, search for the gene by name. In this example, we'll search for evidence that the SIN3A gene has a role in type 2 diabetes (T2D).
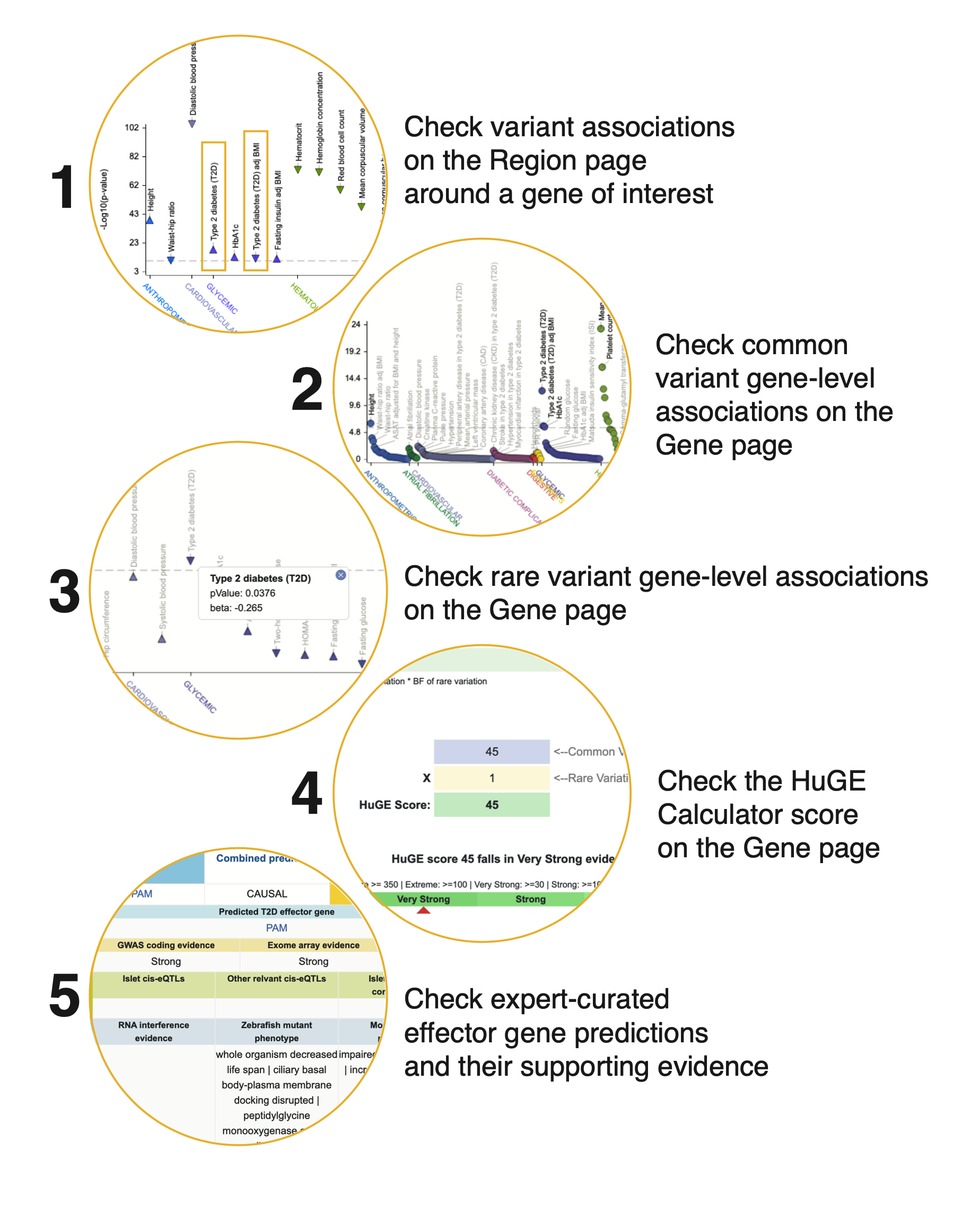
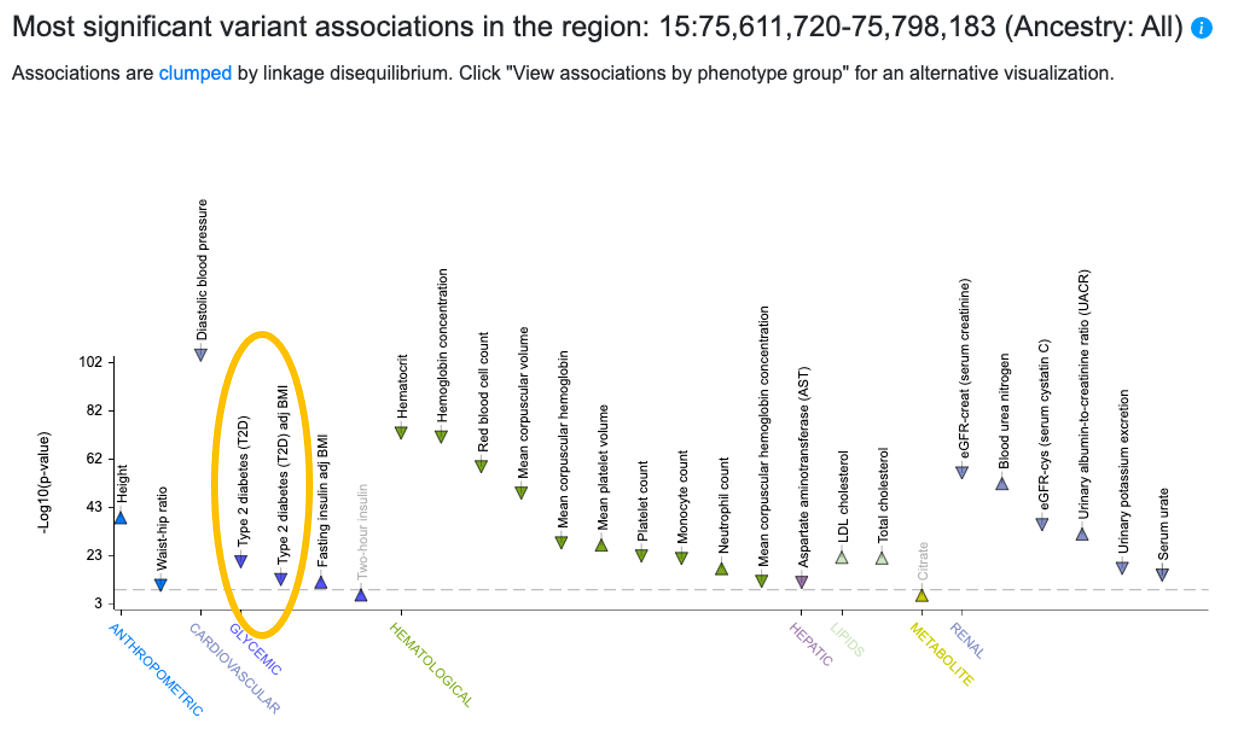
Step 1. Check variant associations on the Region page. Searching by gene name from the home page takes you to the Region page (see complete documentation for the Region page), spanning the coding sequence of the gene and 50 kb of upstream and downstream flanking sequences. On the Region page, check whether there are associations in this region for the phenotype of interest or for related phenotypes. If there is human genetic support for a role of the gene in a phenotype, there will usually be at least one genome-wide significant variant association (p < 5e-8) for that phenotype on the Region page.
The PheWAS plot near the top of the SIN3A Region page shows genome-wide significant associations in the region. We see that there are genome-wide significant associations for T2D and T2D adjusted for BMI.
Step 2. Navigate to the Gene page. To pursue this, let's look at the Gene page for SIN3A. Click on a blue box just above the Region page PheWAS plot to navigate to a Gene page.
Step 3. Check common variant gene-level associations. Near the top of the Gene page for SIN3A, click the Common variant associations tab to view another PheWAS plot. This one displays gene-level associations for SIN3A across all phenotypes available in the Portal, and the associations are also listed in the table below. We calculate them from common variant genetic associations using the MAGMA algorithm. A generally accepted threshold for significance of MAGMA results is p ≤ 2.5e-6, so we see that the SIN3A association with T2D meets this criterion. However, it should be remembered that although a MAGMA significant result indicates that the gene is close to a significantly associated variant, this does not necessarily mean that the gene itself is causal for the phenotype. If multiple genes are located near to a significantly associated variant, MAGMA assigns low p-values to all of them.
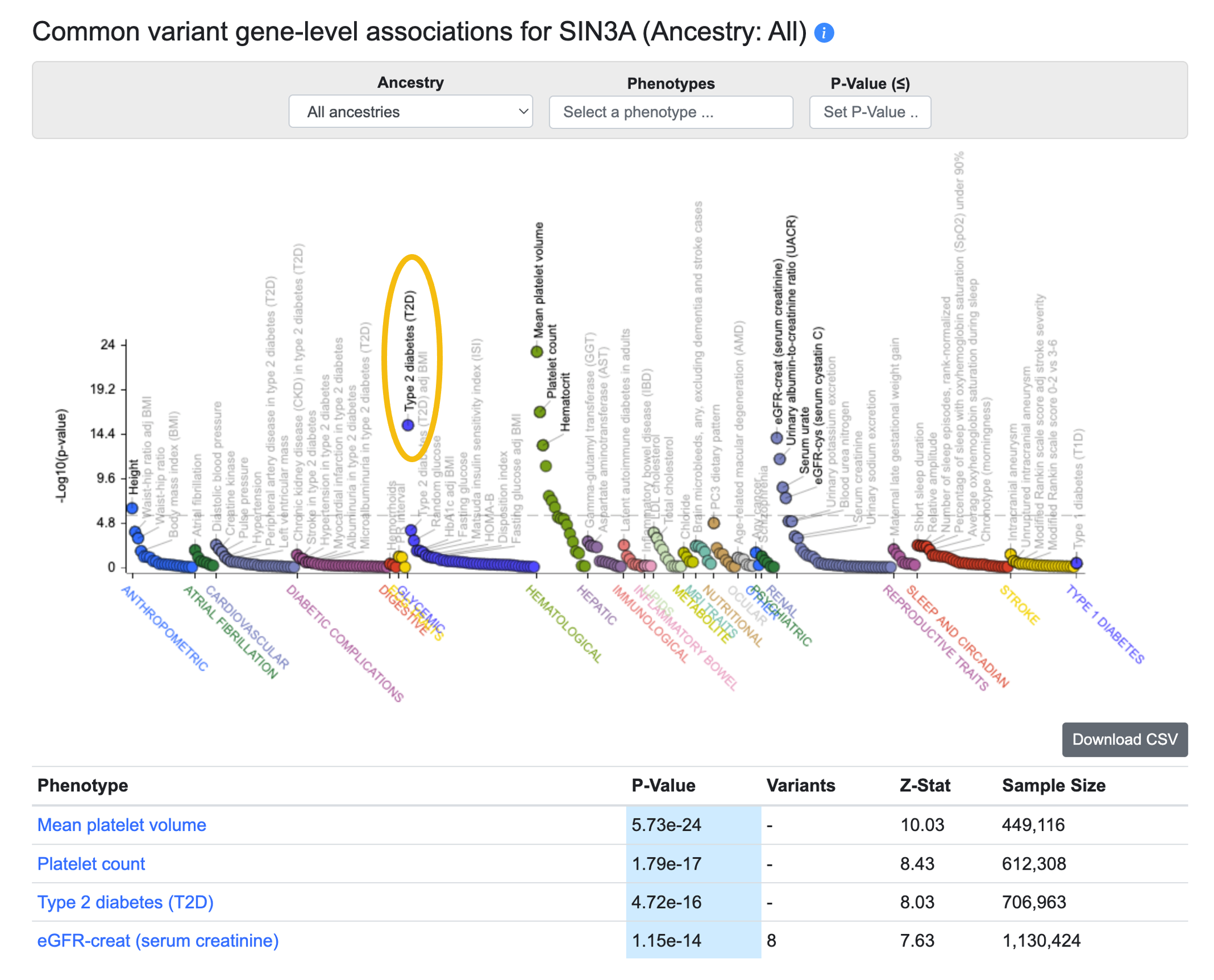
Step 4. Check rare variant gene-level associations. The Rare variant associations tab displays another PheWAS plot with an accompanying table. This one displays gene-level associations calculated from rare variants. For each phenotype and gene, a burden test was performed to calculate a gene-level association score based on the associations of variants within the coding sequence. See the Gene page guide for all the details on this method.
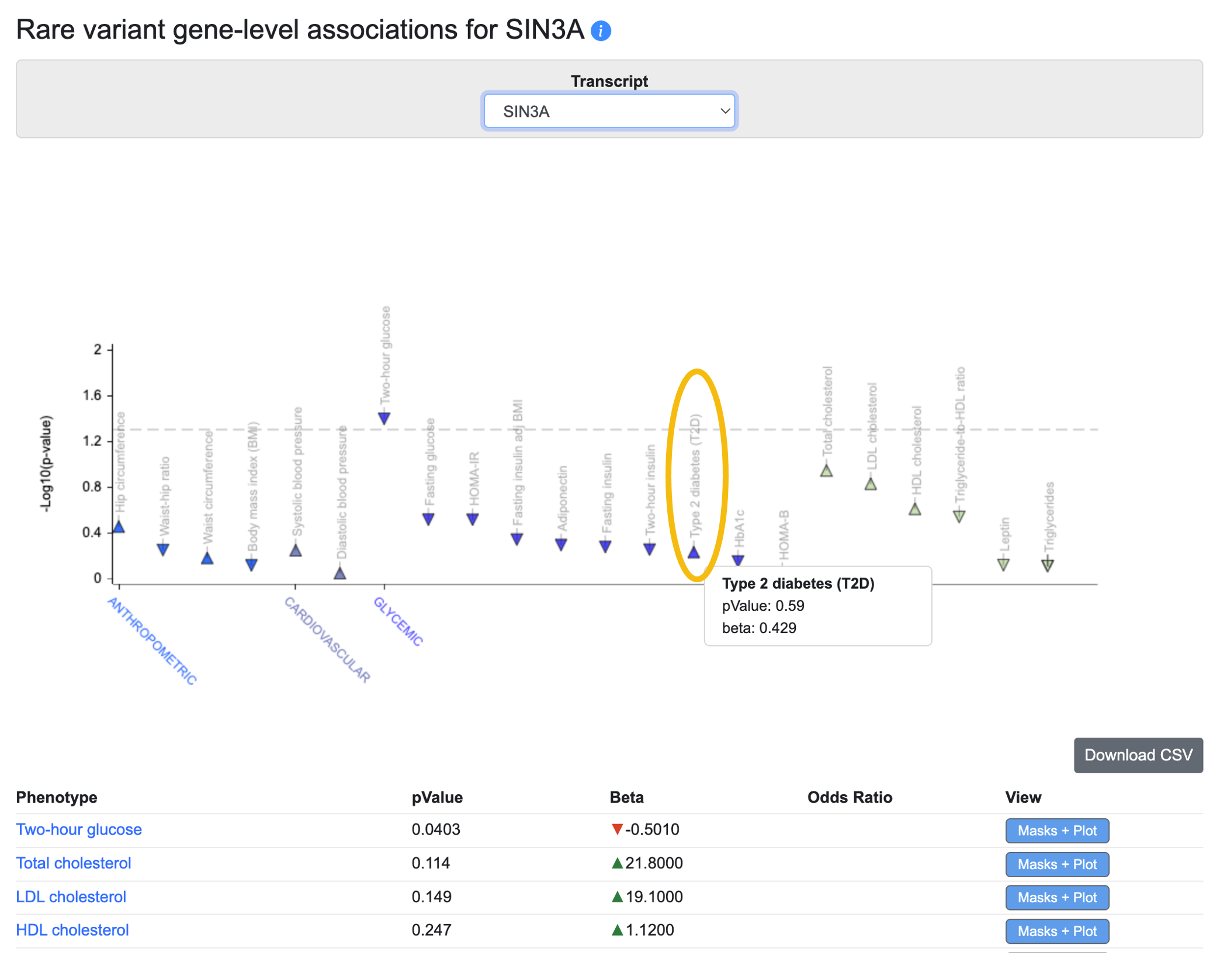
Here we see that the rare variant-based gene-level association for SIN3A with T2D does not meet the generally accepted threshold for exome-wide gene-level significance of p ≤ 6.57 x 10-7.
Step 5. Check the HuGE Calculator score. For a simple summary that takes into account both common and rare variation to evaluate genetic support for the involvement of a gene in a phenotype, click the HuGE Scores tab at the top of the Gene page. Detailed documentation on the HuGE Calculator is available here.

The HuGE Calculator score shows us that, despite the lack of strong rare variant evidence for the involvement of SIN3A in T2D, the overall evidence is "Very strong".
Step 6. Check expert-curated effector gene predictions. The Portal includes predictions generated by researchers who integrate many disease-relevant data types to predict the likely causal genes at genetically associated loci. If there is an effector gene prediction list for your disease or trait of interest, the presence of a gene of interest on the list can be further support for its role. Depending on the methods used to make the prediction, the list may display additional relevant genetic or genomic evidence, beyond the results integrated in the portal.
To see lists of predicted effector genes, navigate to the Predicted Effector Genes link under the KP Labs menu.
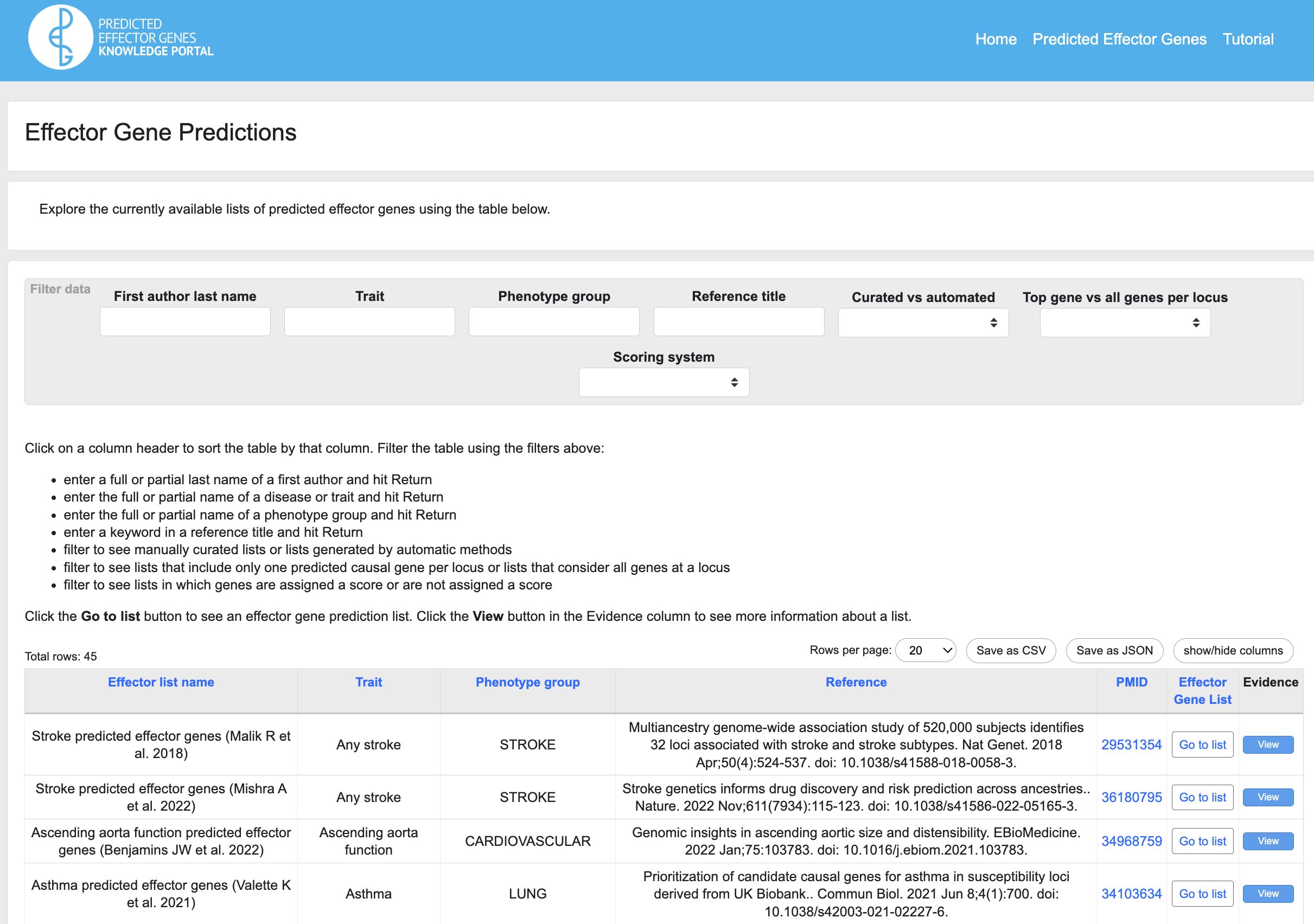
Use the filters to narrow down the table to display predicted effector lists for phenotypes of interest. Click the Go to list button to view a list. The lists can be searched by gene name and other criteria. An "Evidence" button in each row expands the row to show more supporting evidence.
Please contact the Knowledge Portal team if you would like assistance in evaluating the genetic and genomic evidence available in the portals.
