Meta-analysis of genetic association studies provides an estimate of the effect of a genetic variant on a phenotype by pooling information across multiple studies with similar designs and sampling. It can augment our knowledge, by identifying novel associations that are not significant at the level of individual datasets but become significant when multiple studies are considered.
One potential complication in performing meta-analysis is when samples overlap between the studies. Failing to account for this overlap can lead to inflated type 1 error (incorrectly identifying variants as associated with a phenotype when in fact they are not).
Our bottom line analysis estimates the sample overlap between each pair of studies/datasets and accounts for it when weighting each study’s contribution to the final effect estimate. The method is implemented in METAL (documented here) and was developed at the University of Michigan. The procedure for performing this analysis in the Knowledge Portals is outlined below.
Please note that this method as instantiated in the Knowledge Portals is experimental; be sure to compare the results with those from individual datasets. We are happy to provide help in evaluating these results; please contact us.
View this webinar for an in-depth discussion of the bottom-line analysis:
Bottom-line integrative analysis method
Some datasets loaded to the Knowledge Portals are single-ancestry (European (EU), African American (AF), etc.) while others include multiple ancestries (Mixed):
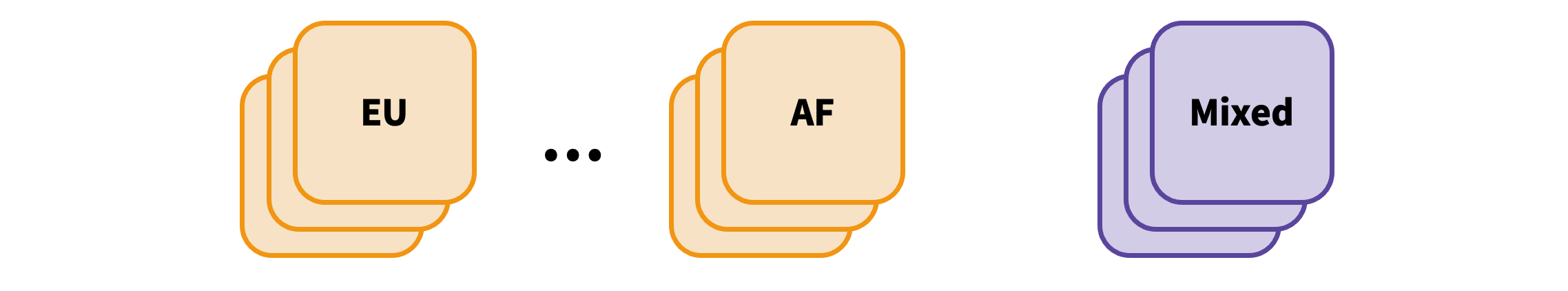
For each dataset, the variants are first filtered to remove multiallelic variants and variants that are missing p-value, beta/OR, or sample size.
The variants in the single-ancestry datasets are then partitioned into common (allele frequency > 5%) and rare (allele frequency ≤ 5%) variants:
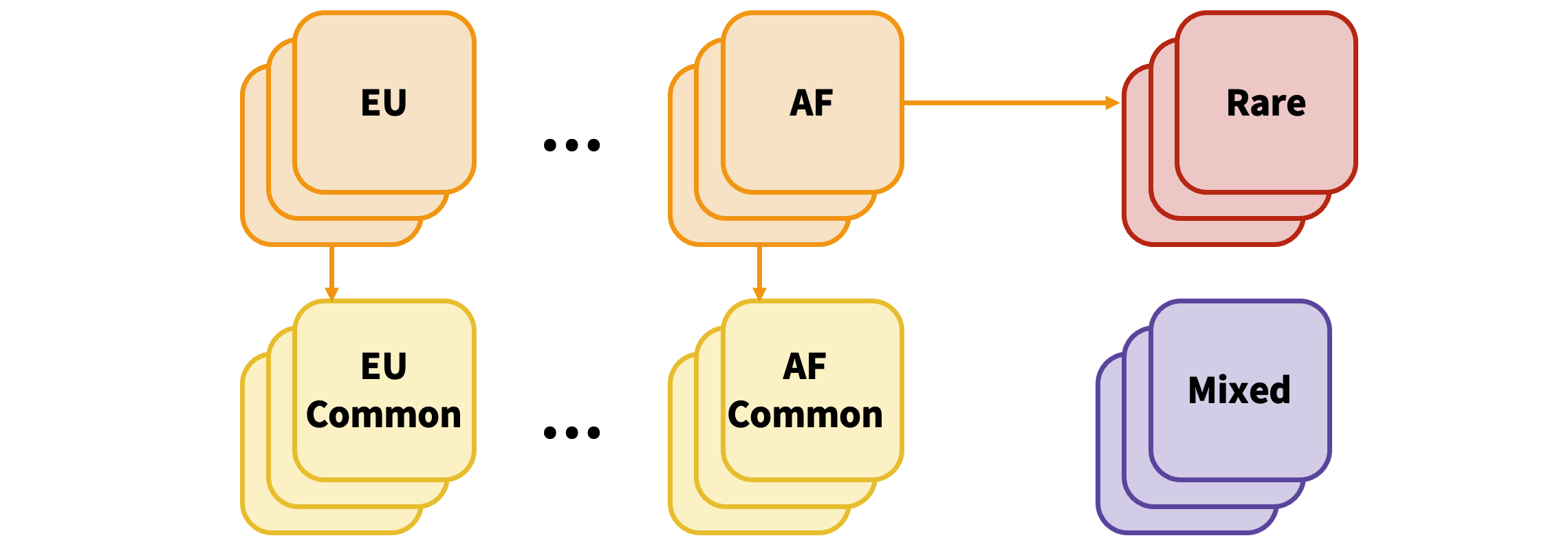
METAL is run over common variants from each single-ancestry dataset with OVERLAP ON:
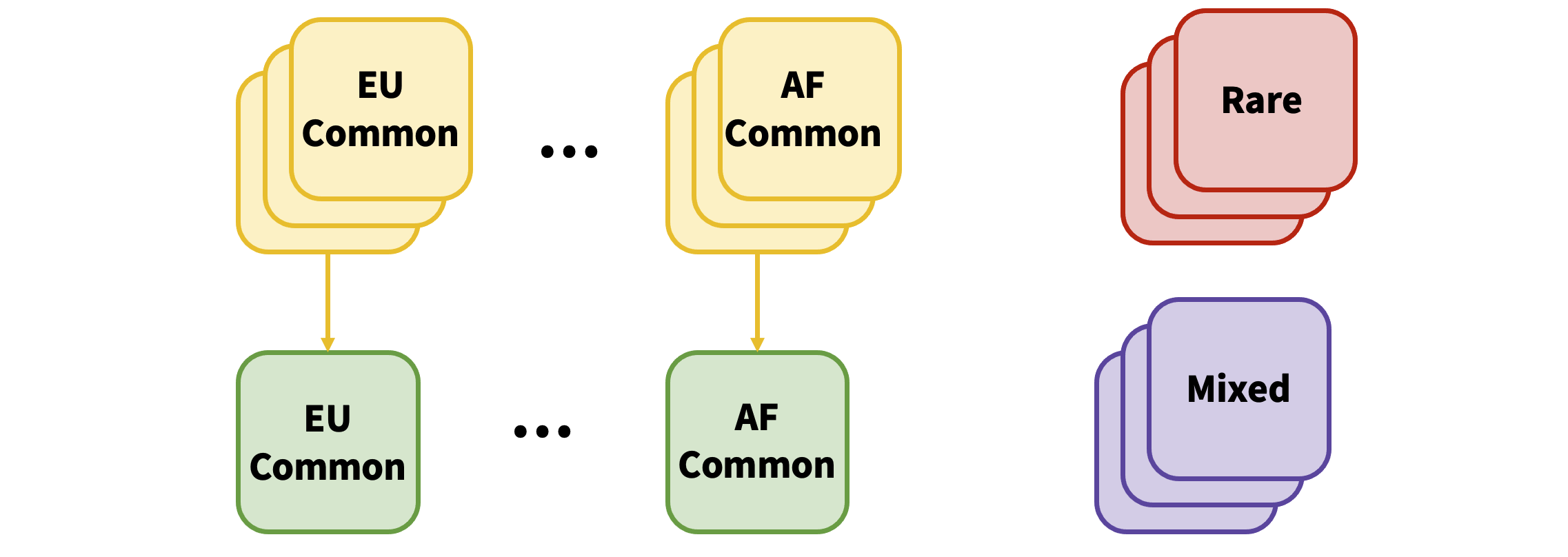
The output is then unioned with the rare variants from all datasets:
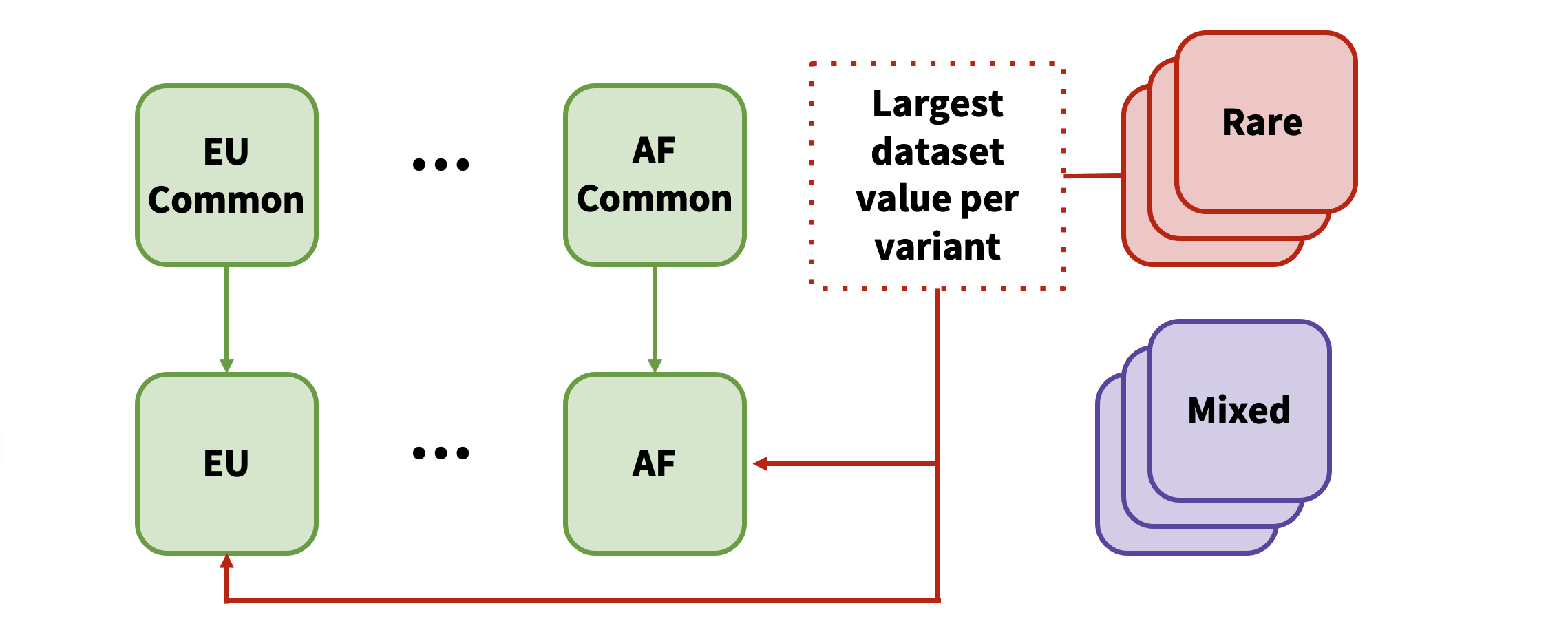
If a rare variant is present in more than one dataset, only the association from the largest dataset is added. In the event that a variant exists as both common and rare (from differing datasets), only the variant with the largest total sample number across all datasets is kept.
After each ancestry has been processed, METAL is run across all the ancestries with OVERLAP OFF:
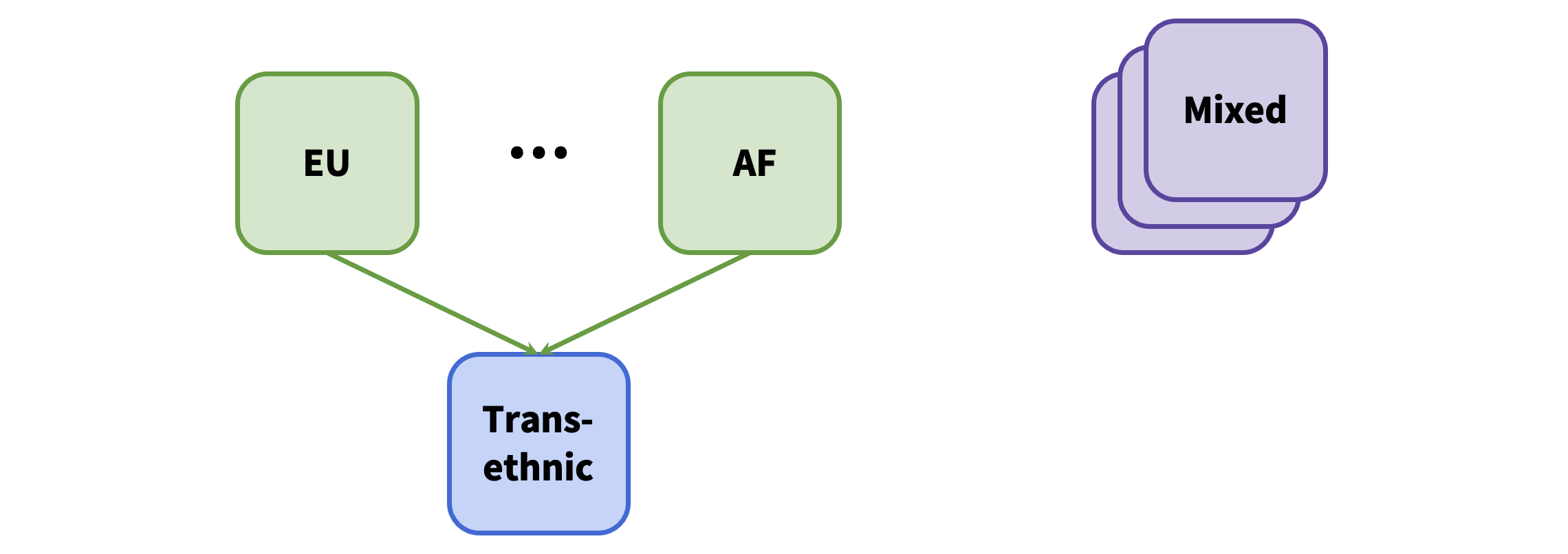
If "Mixed" ancestry datasets were excluded from the ancestry-specific analysis (due to the presence of at least one non-mixed dataset), then mixed ancestry datasets are loaded and the following operations are performed on the trans-ethnic results:
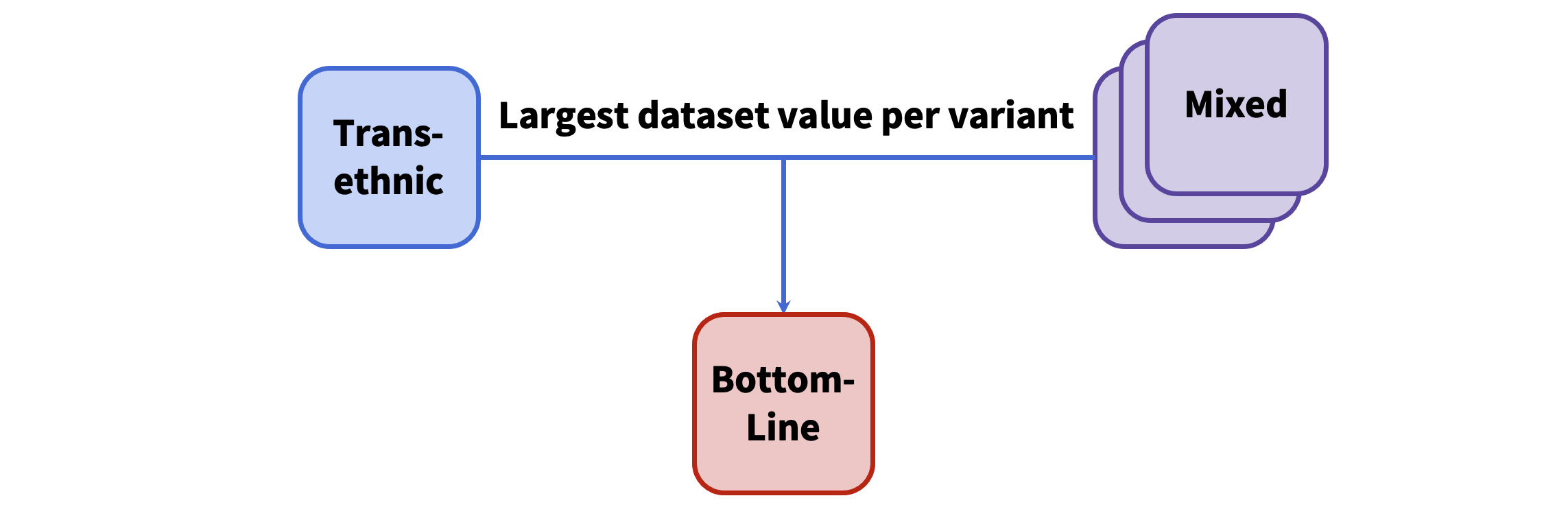
- Any mixed-ancestry variants with a larger, single-dataset sample size than the combined, trans-ethnic sample size will replace the trans-ethnic result.
- All mixed-ancestry variants are added to the trans-ethnic results, and the association from whichever dataset or meta-analysis has the largest sample size is kept.
- The final results of the trans-ethnic analysis are loaded into the database as "bottom line" results.
If no single-ancestry datasets exist for a phenotype, results from mixed-ancestry datasets are simply displayed on the portal.
