Integrating genetic and genomic results to predict which genes are effectors for a disease or trait can help researchers to prioritize further experimentation and to better understand disease mechanisms and pathways. To facilitate these goals, we are aggregating effector gene predictions for common metabolic diseases in the CMDKP. Today, we are making available predictions for coronary artery disease (CAD) effectors from a new study by members of the CARDIoGRAMplusC4D consortium (Aragam et al., medRxiv, 2021).
You can access all available predictions on the CMDKP Effector gene predictions page, which is reached via the "Tools" menu in the upper menu bar on every CMDKP page. The Effector gene predictions page lists, describes, and links to the currently available results.
The CARDIoGRAMplusC4D CAD effector gene predictions are based on a large new GWAS conducted by the consortium that analyzed CAD associations in more than 180,000 cases and nearly a million controls. After fine-mapping the signals, the researchers applied 8 prediction methods to the genes in the associated loci to generate a list of 185 potential effector genes. To get a quick overview of the study, watch this brief video "explainer" created by Jeanette Erdmann.
The list of these candidate genes is displayed in an interactive interface on the CMDKP along with details of the evidence supporting the predictions. At the top of the page is a Manhattan-like plot of the chromosomal location for the sentinel variant in each GWAS locus versus the number of predictors for the potential effector gene at that locus. Mousing over a point opens a tooltip with identifying information.
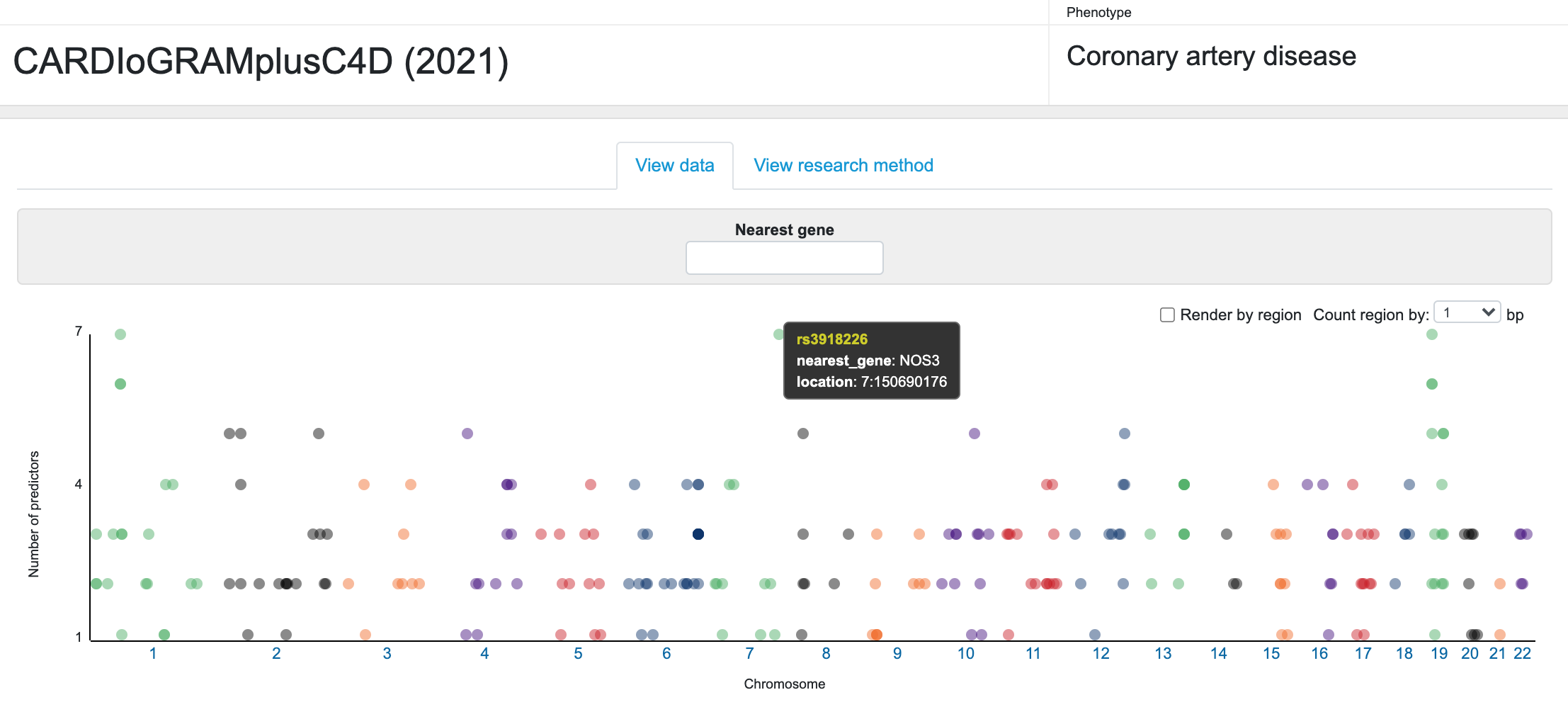
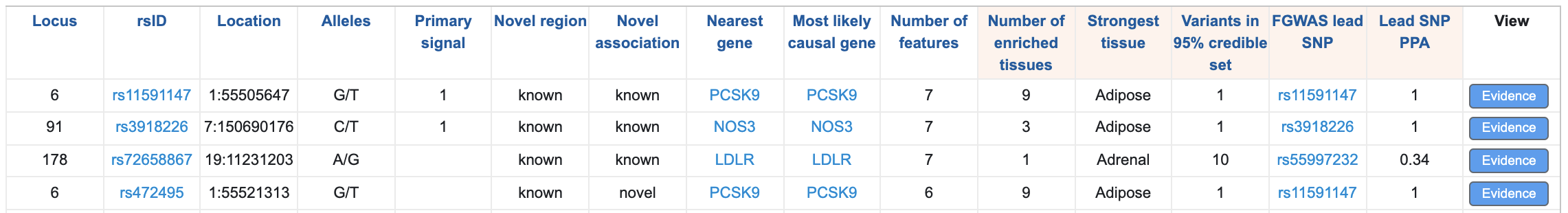
Below the plot, a table lists each locus and displays the supporting evidence. You can filter both the plot and the table to display information for one or more specific genes by typing gene names into the "Nearest gene" box above the plot. The default view of the table summarizes the prediction results for each locus. Click on any column header to sort the table by that column.
Clicking the blue "Evidence" button opens a summary of the detailed evidence for that locus:
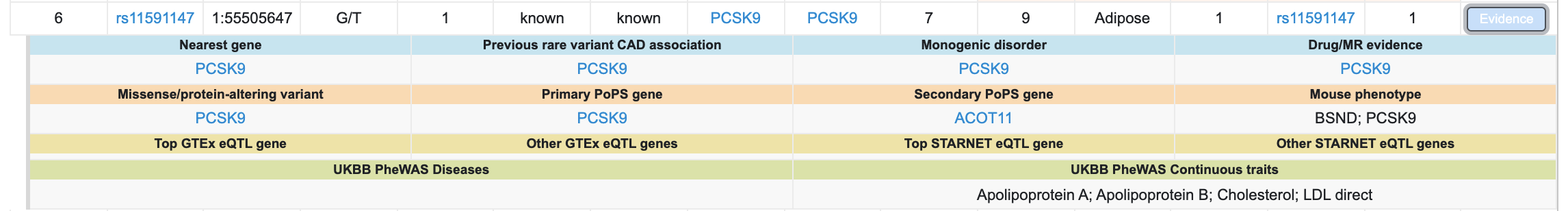
Find further details on the "Research method" tab of the interface or in the preprint. Please give this interface a try and contact us with any questions or suggestions!
