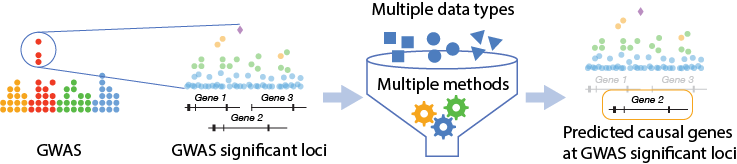
Outline of effector gene prediction
The aim of the working group is to define recommendations for PEG list reporting.
The working group has recently developed the PEGASUS Framework, a FAIR community standard for reporting predicted effector genes derived from GWAS and integrative evidence. We invite community members to benchmark your own results using these standards and share your experience.
For context and general recommendations take a look at our recent publication [Realizing the promise of genome-wide association studies for effector gene prediction, Costanzo et al]. To join the working group, please contact us.
Working Group Meetings
October 7th, 2025
Meeting Notes | Watch working group recording
- Run through of ASHG ancillary session
- Laura - landscape
- Aoife - framework
- Matt - benchmarking
- Laura - discussion guiding questions
September 4th, 2025
Meeting Notes | Watch working group recording
- Introduction & promoting/planning for ASHG Ancillary Session - Julie Jurgens
- Summary of benchmarking activity - Julie Jurgens
- Presentation of benchmarking results - Benchmarking Team
- Fitting PEG matrix standards to PEG lists - Laura Harris/Aoife McMahon
July 9th, 2025
Meeting Notes | Watch working group recording
- Update on benchmarking activity - Aoife McMahon
- Discuss based on review of template - Aoife McMahon
- High-level metadata intro, considerations (e.g., AI/ML readiness, ontologies) - Aoife McMahon
- Metadata standards - Marcos Casado Barbero
- Our attempts at abstraction of metadata schema - Yue Ji
- Discussion: Metadata evidence categories and their suitability, informed by benchmarking - all
June 5th, 2025
Meeting Notes | Watch working group recording
- Working Group Roadmap: ASHG & Beyond - Julie Jurgens
- Data Content Standards - Aoife McMahon
- Assigning Benchmarking Activity - Yue Ji & Noel Burtt
May 8th, 2025
Meeting Notes | Watch working group recording
- Reflections - Maria Costanzo
- Recap of September 2024 Predicted Effector Gene Workshop - Aoife McMahon
- Introduction of the WG - Julie Jurgens
- Moving from guidelines to standards for PEG data - Laura Harris
Working Group Members
| Name | Institution |
|---|---|
| Zhanna Balkhiyarova | University of Surrey |
| Marcos Casado Barbero | EMBL-EBI |
| Oleg Borisov | University of Freiburg |
| MacKenzie Brandes | Broad Institute |
| Noel Burtt | Broad Institute |
| Adam Butterworth | University of Cambridge |
| Nathalie Chami | Icahn School of Medicine at Mount Sinai |
| Daniel Considine | Open Targets Genetics |
| Maria Costanzo | Broad Institute |
| Ayse Demirkan | University of Surrey |
| Eric Fauman | Pfizer |
| Xiangyu Ge | Open Targets Genetics |
| Laura Harris | EMBL-EBI |
| Karl Heilbron | Charité Universitätsmedizin Berlin |
| Quy Hoang | Broad Institute |
| Yue Ji | EMBL-EBI |
| Julie Jurgens | Broad Institute |
| Emrah Kacar | Trinity College Dublin |
| Kanika Kanchan | NIAID/NIH |
| Lillya Kopanitsa | Wellcome Sanger Institute |
| Yong Li | University of Freiburg |
| Aoife McMahon | EMBL-EBI |
| Ellen McDonagh | EMBL-EBI/Open Targets Genetics |
| Nina Oparina | Uppsala University |
| Matt Pahl | Children's Hospital of Philadelphia |
| Santhi Ramachandran | EMBL-EBI |
| Wafaa Rashed | Ahram Canadian University-Egypt |
| Gabi Rinck | EMBL-EBI |
| Oliver Ruebenacker | Broad Institute |
| Abdurrahman Shiyanbola | University of Surrey |
| Loz Southam | Institute of Translational Genomics, Helmholtz Munich |
| Szymon Szyszkowski | Open Targets Genetics |
| Sylvanus Toikumo | University of Pennsylvania |
| Yakov Tsepilov | Wellcome Sanger Institute |
| Kyle Vogan | Nature Genetics |
| Sean Yao | Insilico Biosystems |
| Juliana Xavier de Miranda Cerqueira | University of Tampere |
| Chi Zhang | ByteDance |
| Norann Zaghloul | NIDDK/NIH |
First community workshop on standards and infrastructure for predicted effector gene lists
Held September 16-17, 2024 | In-person at the Broad Institute, Cambridge MA USA | In-person at the European Bioinformatics Institute, Hinxton, UK | By videoconference
Researchers who perform genome-wide association studies (GWAS) often aggregate and integrate multiple other evidence types in order to predict the effector (causal) genes at GWAS loci (see some examples). These predictions are a valuable outcome of GWAS, but in 2024 there were currently no community standards for organizing or sharing them, and the proliferation of multiple evidence types, methods, and presentation formats carried the risk of confusion. To spark discussion on how to address this gap, we held a community workshop on the standards, infrastructure and incentives required to promote and enable sharing, interoperability, and updating of predicted effector gene lists. In preparation for the workshop, we conducted a community survey about effector gene prediction (see survey results).
The workshop had more than 80 participants from diverse institutions and career stages. A manuscript describing its proceedings is in preparation.
Organizing committee of the September 2024 workshop

Aims of the workshop
- Raise awareness of the diversity of approaches to creating predicted effector gene (PEG) lists
- Identify challenges in sharing, maintaining, and updating PEG lists
- Ensure that PEG lists with their supporting evidence and related metadata are FAIR and relevant to the user community
- Begin to discuss a community standard for reporting PEG lists
- Define strategies for evaluation and collation of PEG lists
Identify strategies for incentivizing implementation/use of the proposed standards
Agenda
Monday September 16, 2024: 9am - 1pm EDT / 2pm - 6pm BST | View video recording
- 9:00 EDT / 2:00 BST: Introduction and welcome (Noël Burtt, Knowledge Portal Network)
- 9:20 EDT / 2:20 BST: Who’s that causal gene? Lessons learned in manually annotating 100,000 GWAS SNPs (Eric Fauman, Pfizer Inc.)
- 10:20 EDT / 3:20 BST: Open Targets as an end user of PEG lists (Yakov Tsepilov, Open Targets)
- 10:35 EDT / 3:35 BST: Open discussion
- 10:45 EDT / 3:45 BST: Break
- 10:55 EDT / 3:55 BST: Overview of current gene prioritization efforts and PEG lists (Maria Costanzo, Knowledge Portal Network)
- 11:25 EDT / 4:25 BST: 10-minute presentations of gene prioritizations
- Cassandra Spracklen, University of Massachusetts
- Brent Richards, McGill University
- Adam Butterworth, University of Cambridge
- 12:05 EDT / 5:05 BST: Open discussion: what are the key features of each approach and how do they compare?
- 12:30 EDT / 5:30 BST: How ClinGen handles framework standardization with input from many invested users (Marina DiStefano, ClinGen)
- 1:00 EDT / 6:00 BST: Adjourn
Tuesday September 17, 2024: 9am - 1pm EDT / 2pm - 6pm BST | View video recording
- 9:00 EDT / 2:00 BST: GWAS Catalog’s experience developing GWAS standards (Laura Harris, GWAS Catalog)
- 9:15 EDT / 2:15 BST: Curation and display of PEG lists in the Knowledge Portals (Maria Costanzo, Knowledge Portal Network)
- 9:25 EDT / 2:25 BST: Curation and display of gold standard lists at Open Targets (Xiangyu Jack Ge, Open Targets)
- 9:35 EDT / 2:35 BST: Open discussion: advantages/disadvantages of PEG list presentations
- 9:50 EDT / 2:50 BST: Results of community survey (Maria Costanzo, Knowledge Portal Network)
- 10:00 EDT / 3:00 BST: Standardization requirements for use of PEG lists as input for computational and AI methods (Jason Flannick, Boston Children’s Hospital)
- 10:15 EDT / 3:15 BST: Break
- 10:25 EDT / 3:25 BST: Proposal for PEG list standards (Yue Ji and Laura Harris, GWAS Catalog)
- 10:40 EDT / 3:40 BST: 40-minute breakout group discussions about the proposed standards. Guiding questions will be provided.
- 11:20 EDT / 4:20 BST: Breakout groups report back; further discussion in the larger group
- 12:40 EDT / 5:40 BST: Wrap up (All organizers)
- 1:00 EDT / 6:00 BST: Adjourn
