In the Knowledge Portal Aggregator we use the method of LD score regression (Finucane et al., 2015; Bulik-Sullivan et al., 2015; GitHub repository) in two calculations.
We use cross-trait LD score regression (LDSC) to calculate genetic correlations between phenotypes. These are displayed on Phenotype pages (see an example).
We apply stratified LD score regression (S-LDSC) to the bottom-line ancestry-specific genetic associations in the Knowledge Portal database to calculate global enrichment of genetic associations within tissue-specific epigenomically annotated regions such as enhancers, promoters, open chromatin, etc. Significant enrichment of genetic association signals for a disease in annotated regions of a particular type in a specific tissue suggests that the tissue may be relevant for the disease. We previously used the GREGOR method for this calculation, but have replaced it with S-LDSC because S-LDSC is more robust to potential confounders and produces more accurate and specific enrichments.
Globally enriched annotations are displayed on Phenotype pages (see an example) and on the Variant Sifter interface.
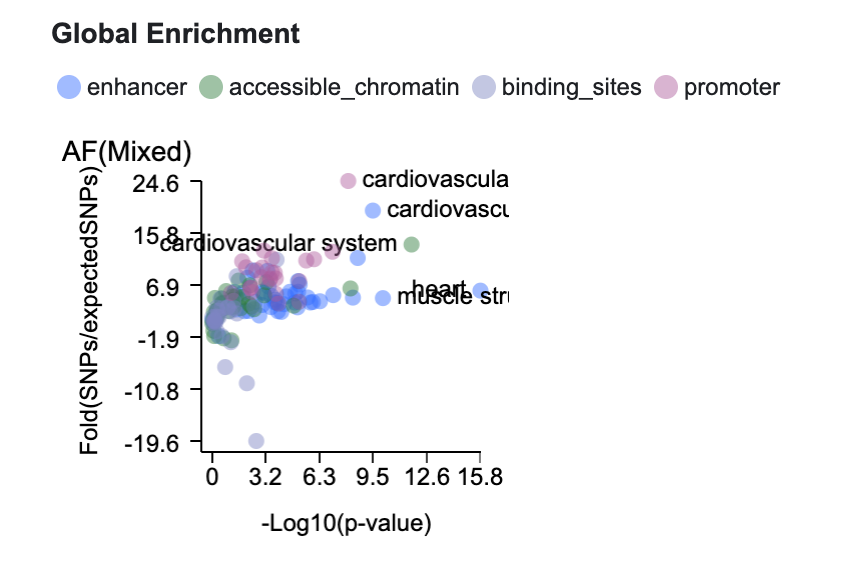
As an example, this global enrichment plot on the Variant Sifter interface representing the values generated by S-LDSC suggests that atrial fibrillation genetic associations are most significantly enriched in regions annotated as promoters, enhancers, and accessible chromatin in cardiovascular tissues.
