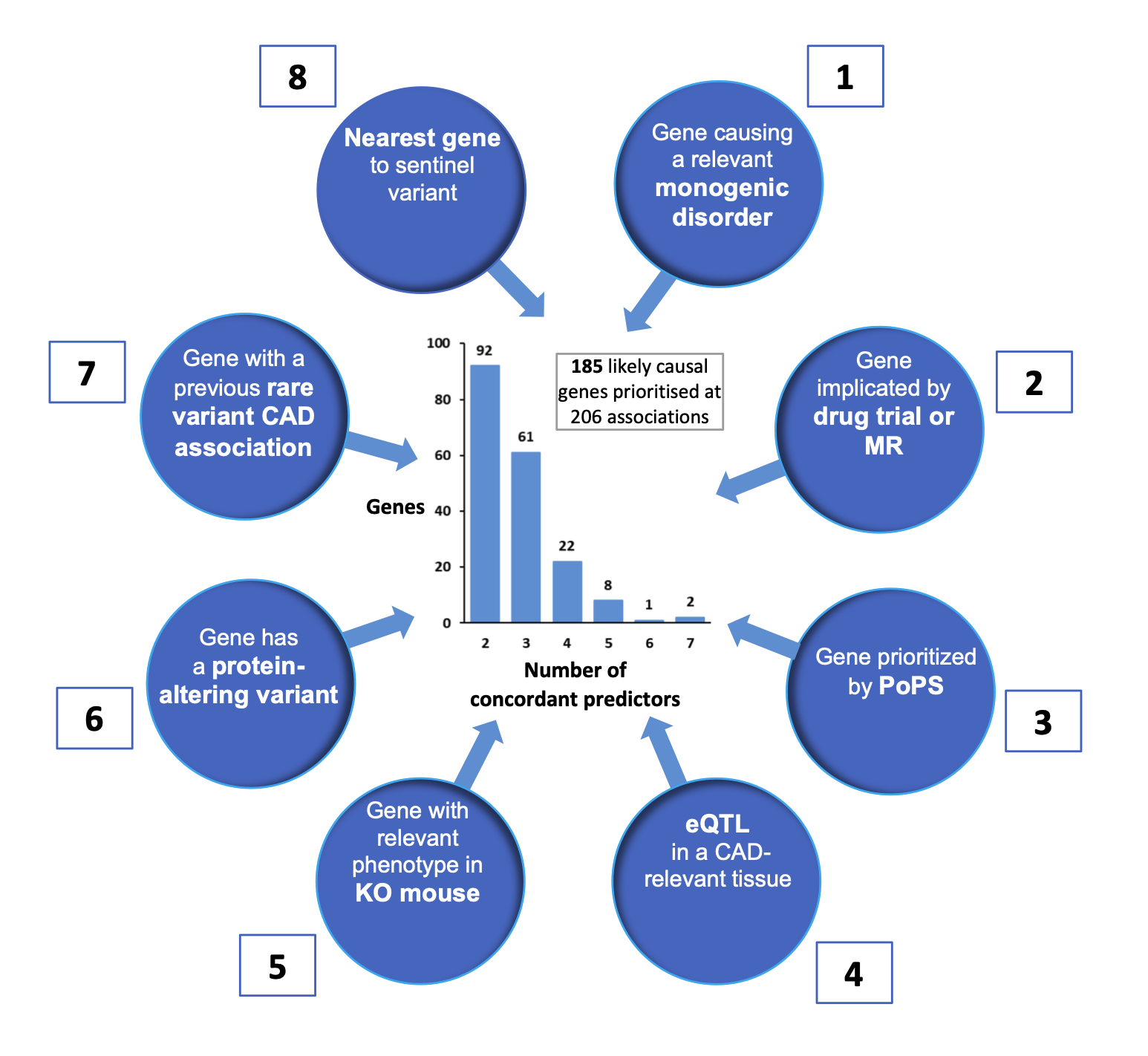
To prioritize likely causal genes for each of 241 genomewide-significant CAD association signals, eight locus-based or similarity-based predictors of causal genes were considered:
- Genes harboring pathogenic (or likely pathogenic) variants responsible for monogenic disorders of cardiovascular relevance according to ClinVar
- Genes encoding proteins of established causal relevance to CAD per Mendelian randomization studies, or that are targets for established cardiovascular drugs;
- The two genes with the highest Polygenic Priority Score (PoPS) within 500kb of the sentinel variant;
- Genes with eQTLs in CAD-relevant tissues from STARNET or GTEx
- Genes that display cardiovascular-relevant phenotypes in knock-out mice from the International Mouse Phenotyping Consortium or the Mouse Genome Informatics database
- Genes containing protein-altering (i.e. missense or predicted loss-of-function) variants that are in strong LD (r2≥0.8) with the CAD sentinel variant;
- Genes containing rare coding variants that have been associated with CAD risk in previous whole-exome sequencing or array-based studies;
- The nearest gene to the CAD sentinel variant.
See the figure below for a summary. Genes were prioritized as "likely causal" for each sentinel association using a consensus-based approach, selecting the gene with the highest, unweighted sum (0-8) of evidence across all eight predictors. Most likely causal genes were assigned for 206 associations where there were at least two concordant predictors for a gene, prioritizing 185 distinct genes. 94 of these genes had three or more concordant predictors, so were considered "strongly prioritized".
Further details can be found in Aragam et al., medRxiv, 2021. Watch an "explainer" video about this study, created by Jeanette Erdmann.