The Effector Index (Ei) algorithm calculates the probability of causality for each gene at a locus that harbors genome-wide significant SNVs for a disease or trait.
Full documentation is available in:
An effector index to predict target genes at GWAS loci.
Forgetta V, Jiang L, Vulpescu NA, et al.
Hum Genet. 2022 Feb 11. doi: 10.1007/s00439-022-02434-z.
PMID: 35147782
Development of the Ei predictive model
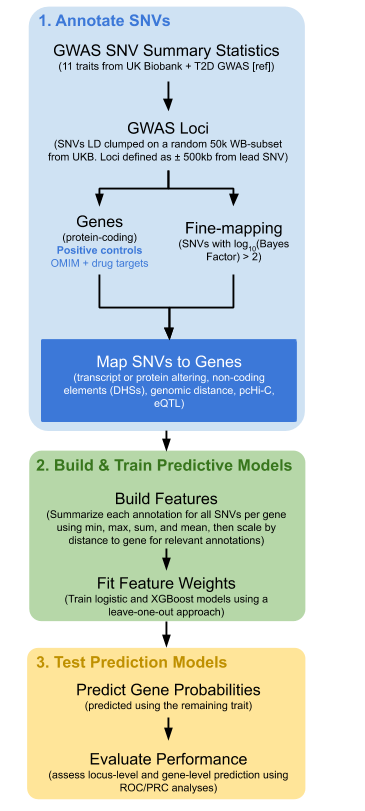
1. Annotate single-nucleotide variants (SNVs). A set of positive control genes was identified by including genes that either encode a protein which is the target for a medication used in humans to treat the disease or trait, or the gene causes a Mendelian form of the disease (or influences the trait). Second, for each trait of interest, GWAS results were obtained and significant loci identified (defined as P ≤ 5 ×10-8). Third, at each locus, we undertook statistical fine-mapping and mapped putative causal SNVs to genes, by annotating the SNVs for putative biological function, such as altering the transcript or protein, association with gene expression (eQTL), or intersection with non-coding elements (DHSs). Annotations were assessed for their ability to enrich for positive control genes. Each annotation was then summarized for all SNVs per gene using min, max, sum, and mean, followed by scaling by distance to gene for relevant annotations.
2. Build and train predictive models. The SNV annotations were then used for model building and testing. Using only loci known to contain at least one positive control gene, predictive models were built using logistic regression and XGBoost.
3. Test prediction models. We tested the performance of the models on traits and genes not included in the model building. Measurement of model performance was assessed using a leave-one-out approach, wherein the model was built iteratively using eleven traits and tested on the twelfth. To further assess model performance, a model training was generated on all traits and diseases except T2D. This model was then tested on an independently curated set of positive control genes for T2D, which included drug targets, OMIM genes and genes identified by large-scale exome sequencing and exome arrays. The effector index was further tested against simpler approaches, including the gene nearest the lead SNV.
