In our latest release, we've added four new or updated items to our "KP Labs" menu. KP (Knowledge Portal) Labs (named with a nod to the Google incubator, Google Labs), is a space where we offer beta versions of tools and interfaces that are in development. We're confident that the underlying data are solid, but the user interfaces may not yet present optimal workflows, and yet-undiscovered bugs could even be present. We release these tools in the hopes that you, the Knowledge Portal users, will try them out and let us know what you like or don't like, what you find useful, and what improvements you'd like to see.
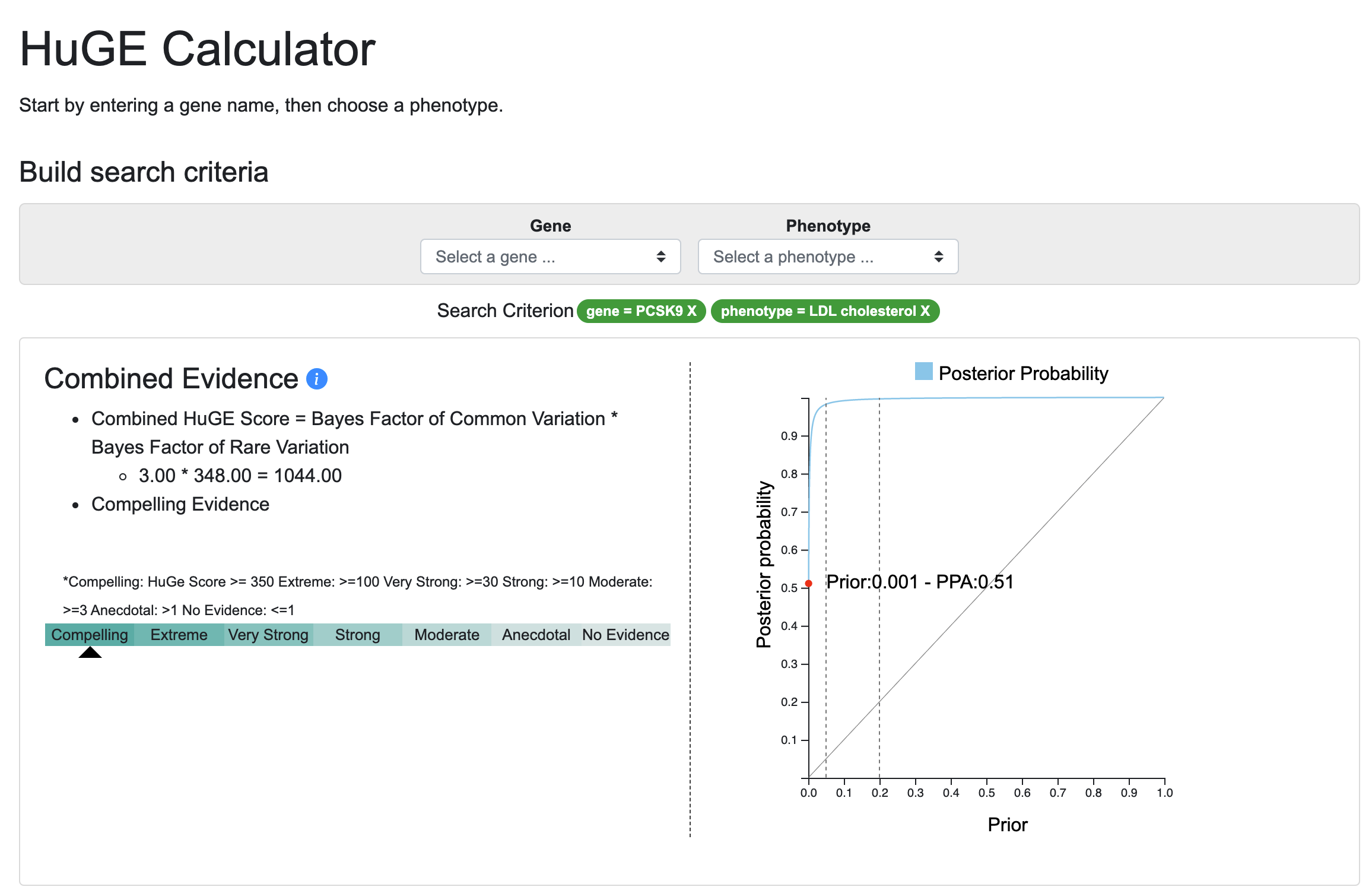
Judging from the 100 attendees at the webinar describing it, and more than 100 views of the webinar recording, there's a lot of interest in the Human Genetic Evidence (HuGE) Calculator. Developed by Peter Dornbos and Jason Flannick, this tool offers a way to quantify genetic evidence supporting involvement of a gene of interest in a disease or trait. We present the HuGE Calculator in the KP Labs space ahead of publication.
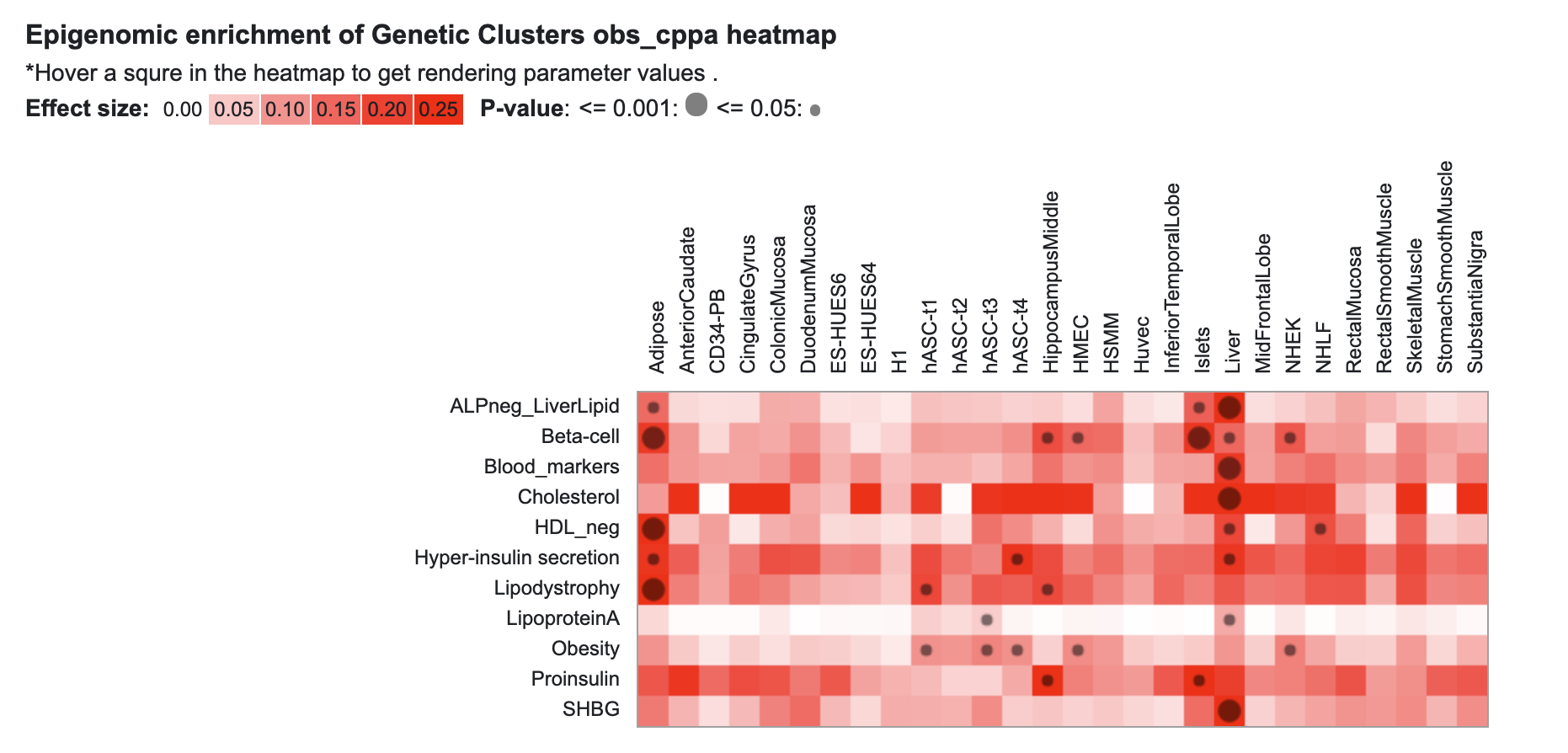
Also pre-publication, KP Labs now includes an update of genetic clustering results for several traits. Previously, we displayed an interface allowing you to view and explore results from a clustering analysis of 94 T2D-associated loci (Udler et al., 2018). That interface is still available, and we've now added results from an updated analysis by Claire Kim and Miriam Udler for loci associated with T2D, coronary artery disease, and chronic kidney disease. Accompanying these results is a heatmap display of tissue-specific epigenomic enrichment in those clusters, generated by Joshua Chiou and Kyle Gaulton.
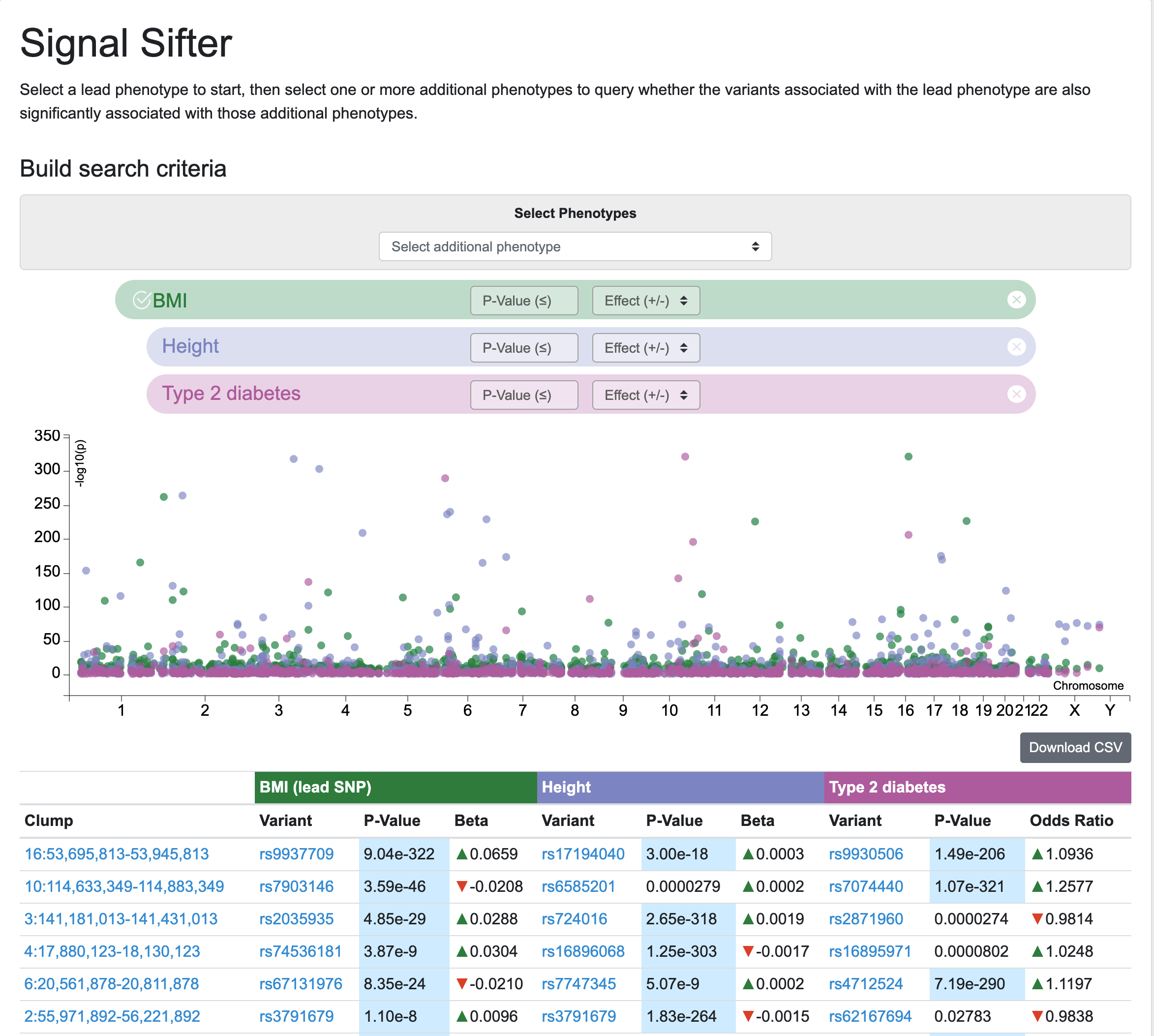
Longtime users of the Knowledge Portals may remember the Variant Finder tool, which retrieved variants associated with multiple phenotypes according to custom criteria. The Variant Finder has now been re-worked as the Signal Sifter, which retrieves LD clumped associations for a lead phenotype and then searches within those clumps for variants associated with one or more secondary phenotypes of choice. Results may be browsed via a table and plot, and are also downloadable.
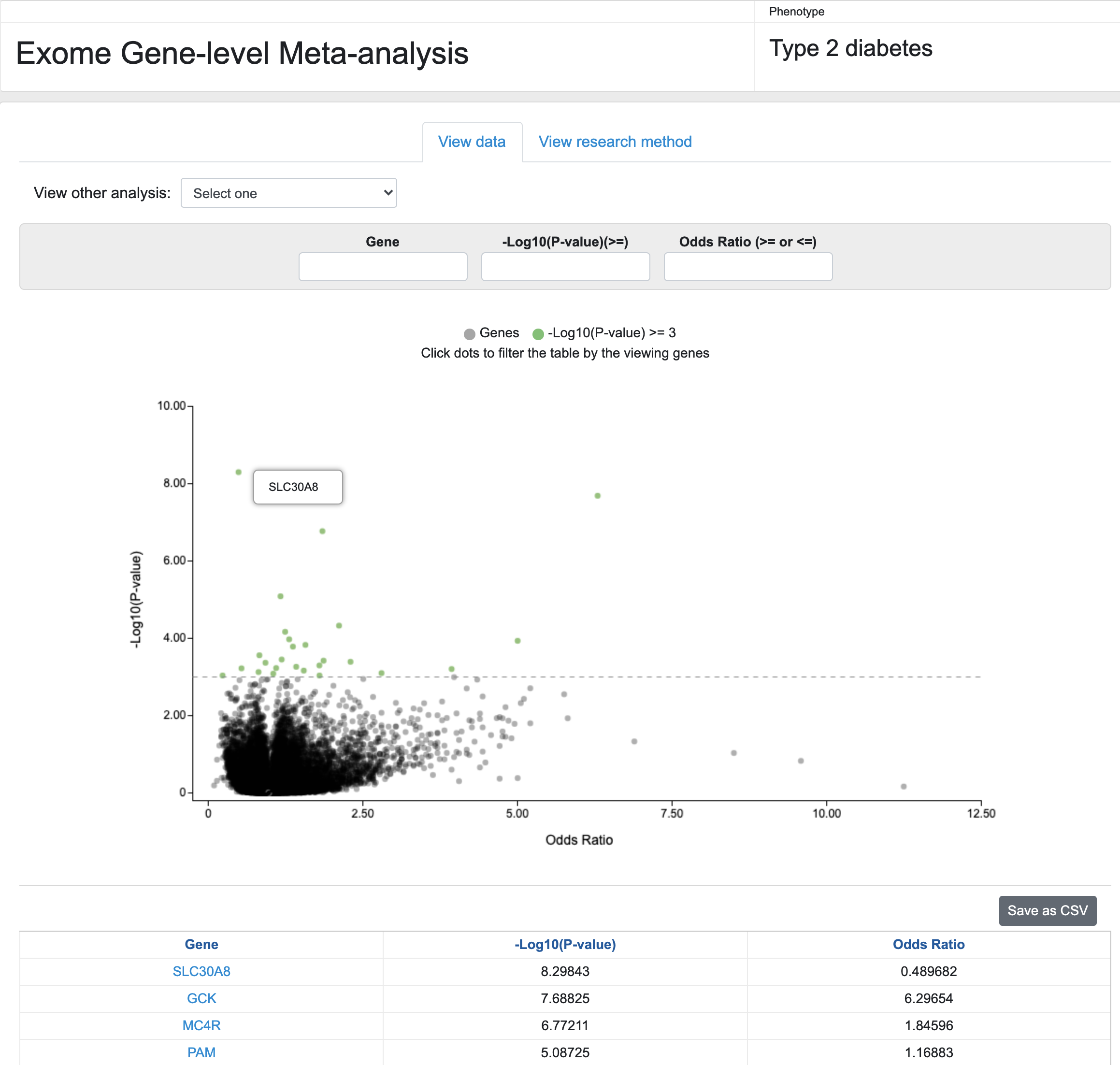
Finally, KP Labs now presents preliminary results from a meta-analysis, performed in Jason Flannick's group, of whole-exome sequencing data from 86,000 samples to generate gene-level association scores for T2D. The interface allows you to select any of 7 different variant masks and then search and explore the results. We present these results with the caveat that although we believe them to be accurate, they are currently not complete and should be analyzed with caution. They produce weaker associations for known positive controls, and investigation of the reason for this is ongoing.
All of the KP Labs tools are available in the Common Metabolic Diseases Knowledge Portal (CMDKP); specific tools are also available in disease-specific portals if their underlying data are relevant to that disease. We would really appreciate your contacting us with input on the KP Labs tools or any other aspect of the Knowledge Portals!
